Abstract
Background:
A subset of human hepatocellular carcinomas (HCC) exhibit mutations of β-catenin gene CTNNB1 and overexpress Glutamine synthetase (GS). The CTNNB1-mutated HCC cell line HepG2 is sensitive to glutamine starvation induced in vitro with the antileukemic drug Crisantaspase and the GS inhibitor methionine-L-sulfoximine (MSO).
Methods:
Immunodeficient mice with subcutaneous xenografts of the CTNNB1-mutated HCC cell lines HepG2 and HC-AFW1 were treated with Crisantaspase and/or MSO, and tumour growth was monitored. At the end of treatment, tumour weight and histology were assessed. Serum and tissue amino acids were determined by HPLC. Gene and protein expression were estimated with RT-PCR and western blot and GS activity with a colorimetric method. mTOR activity was evaluated from the phosphorylation of p70S6K1.
Results:
Crisantaspase and MSO depleted serum glutamine, lowered glutamine in liver and tumour tissue, and inhibited liver GS activity. HepG2 tumour growth was significantly reduced by either Crisantaspase or MSO, and completely suppressed by the combined treatment. The combined treatment was also effective against xenografts of the HC-AFW1 cell line, which is Crisantaspase resistant in vitro.
Conclusions:
The combination of Crisantaspase and MSO reduces glutamine supply to CTNNB1-mutated HCC xenografts and hinders their growth.
Similar content being viewed by others
Main
Hepatocellular carcinoma is the third leading cause of cancer death worldwide (Ferlay et al, 2010; El-Serag 2011). High HCC mortality (Shariff et al, 2009; Ferlay et al, 2010) derives from several causes, including underlying chronic liver disease, late diagnosis, poor resectability, frequent local recurrence, and lack of effective anti-tumour drugs.
Hepatocellular carcinoma is a genetically heterogeneous and complex tumour. So far, molecular data have poorly contributed to explain the clinical variability of the disease and have not resulted into prognostic and predictive biomarkers (El-Serag 2011). Several attempts to correlate transcriptome, genotype and phenotype have been performed (Boyault et al, 2007; Hoshida et al, 2008; Roessler et al, 2012). In one of these (Boyault et al, 2007), six HCC subgroups (G1–G6) associated with clinical and genetic characteristics were identified based on unsupervised transcriptome analysis. G1 and G2 tumours had AKT activation and were associated with low (G1) or high (G2) copy number of HBV and overexpression of genes expressed in fetal liver (G1) or mutations in PIK3CA and TP53. G3 tumours were characterised by mutation of TP53, whereas G4 tumours were an heterogeneous subgroup including TCF1-mutated cancers. G5 and G6 were both related to mutations of CTNNB1, the gene that encodes for β-catenin, and (in particular G6) Wnt pathway activation (Boyault et al, 2007). Tumours with activating mutations of CTNNB1 reproducibly segregate in different studies as a group with distinctive clinical and genetic characteristics (Legoix et al, 1999; Chiang et al, 2008; Cieply et al, 2009).
The frequency of CTNNB1 mutations in human HCC ranges from 3% to 44% (White et al, 2012), leading to the synthesis of a transcriptionally active mutant protein that is resistant to proteasomal degradation and accumulates in the nucleus. These tumours show constitutive activation of the Wnt canonical pathway, display distinctive histology (Dal Bello et al, 2010) and overexpress a number of target genes included in the functional category of amino-acid metabolism (Boyault et al, 2007), such as GLUL, which encodes for Glutamine synthetase (GS). Glutamine synthetase is the only enzyme that is able to synthesise L-glutamine (Gln) from L-glutamate and ammonium. Mutations in other upstream components of the Wnt pathway, such as the AXIN1 gene, may have the same functional effect (White et al, 2012).
Interest on amino-acid metabolism in cancer has been recently fuelled by findings, indicating that some tumours require high Gln availability and are severely inhibited by Gln depletion (Wise et al, 2008). Intracellular Gln can be pharmacologically depleted by means of the bacterial enzyme Crisantaspase (L-Asparaginase from Erwinia chrysanthemi, CRIS) and the GS inhibitor methionine-L-sulfoximine (MSO) (Rotoli et al, 2005; Tardito et al, 2007). Previous results from our group indicated that the CTNNB1-mutated HepG2 cell line was the most sensitive to CRIS and MSO in a panel of HCC cell lines (Tardito et al, 2011).
In the present study, we tested the effects of pharmacologically induced Gln depletion in vivo using murine xenografts induced by two different CTNNB1-mutated HCC cell lines.
Materials and methods
Cell culture
The CTNNB1-mutated HCC cell line HepG2, a gift by prof Giovanni Raimondo, Laboratory of Molecular Biology and Hepatology, Department of Internal Medicine, University of Messina, Italy, was grown in low-glucose (1 g l−1) Dulbecco’s modified Eagle’s Medium (DMEM, cat n. ECM0749L, Euroclone, Pero, Italy), supplemented with 10% of fetal bovine serum (FBS, Lonza, Switzerland), 4 mM Gln and antibiotics (100 U ml−1 penicillin, and 100 μg ml−1 streptomycin).
The CTNNB1-mutated paediatric HCCHC-AFW1 cell line (Armeanu-Ebinger et al, 2012) was grown in high-glucose (4.5 g l−1) DMEM supplemented with 10% FBS, 2 mM Gln and antibiotics (100 U ml−1 penicillin, and 100 μg ml−1 streptomycin). All cultures were maintained at 37 °C in an atmosphere of 5% CO2 in air, pH 7.4.
For in vitro experiments, CRIS (E. chrysanthemi L-Asparaginase, Crisantaspase, EUSA Pharma Ltd., Oxford, UK) was used at 1 U ml−1 and MSO at 1 mM. The effect of CRIS and MSO was assessed counting cells with a Coulter Z1 particle counter. Cells were seeded in complete growth medium in 24-well plates (5 × 104 cell per well) and grown for 24 h. Growth medium was then substituted with fresh medium containing the drugs to be tested. After 72 h, cells were trypsinised and cell number was determined.
Animal experiments
In vivo animal studies were performed according to the Guidelines for the welfare and use of animals in cancer research (Workman et al, 2010) and approved by local ethical Committees, as detailed below.
Assessment of drug toxicity and HepG2 xenografts
To assess the toxicity of the different drug regimens, 5-week-old male athymic nude nu/nu mice (Harlan, Udine, Italy) were treated with intraperitoneal (i.p.) injections of CRIS (5 U g−1) and MSO (10 mg kg−1), both in 0.9% NaCl, or vehicle alone; MSO was injected 3 h after CRIS injection. Treatment was performed three times a week for 3 weeks. Mice were killed 3 h after the last injection of MSO, and peripheral blood (PB) and the liver were immediately collected for analysis. After collection, PB was centrifuged for serum extraction, and serum was deep frozen (−80 °C). Serum albumin and Alanine Aminotransferase (Alanine Transaminase, ALT) were determined with a Dimension Vista 1500 apparatus (Siemens Healthcare Italia, Milan, Italy).
HepG2 xenografts were induced by injecting athymic nude nu/nu mice (Harlan) subcutaneously (s.c.) with HepG2 cells (2.5 × 106 cells). After 8 days, mice were i.p. treated with CRIS and MSO, alone or in combination as described above. Tumour growth was monitored for 19 days by external measurement using a caliper, and tumour volume was calculated by applying the ellipsoid equation. Three hours after the last MSO injection, which corresponds to 6 h after CRIS injection, mice were killed, and PB, tumour masses and livers were collected for analysis. After collection, PB was centrifuged and the serum was deep frozen.
These experiments were performed at the Laboratory of Genetic Engineering for the Production of Animal Models (LIGeMa) at the Animal House of the University of Firenze and were approved by the Italian Ministry of Health (document no. 140/2009-B).
HC-AFW1 xenografts
Xenografts were induced by injecting 8-week-old NOD.Cg-Prkdcscid IL2rgtmWjl/Sz (NSG) mice (Charles River, Sulzfeld, Germany) s.c. with 2 × 106 HC-AFW1 cells. After 2 weeks, mice were treated, as described above, with CRIS, MSO or CRIS + MSO. Tumours were inspected every 3 days, and volumes were calculated by applying the ellipsoid equation. Mice were weighted every 3 days. After 10 days of treatment, mice were killed and PB, tumours and livers were explanted for further analysis. These experiments were performed at the Department of Pediatric Surgery and Pediatric Urology, University Children’s Hospital, Tübingen, Germany, and were approved by the local government’s ethical authority for animal experiments (Regierungspraesidium Tuebingen, No. K 9/11), which are in line with the guidelines for the welfare and use of animals in cancer research.
Amino-acid extraction and determination
Amino acids in serum and tissues and GS activity were measured as previously described (Tardito et al, 2011; Chiu et al, 2012) with minor modifications. Samples were de-proteinised with 10% (w/v) 5-sulfosalicylic acid for 30 min on ice. Samples were then centrifuged for 10 min at 12000 g at 4°C and supernatant was mixed with 1 volume of LiOH buffer (pH 2.2). For tissue amino-acid analysis, liver and tumour samples were homogenised in 6.6 mM solution of acetic acid in ethanol with a homogeniser (Ingenieurbüro CAT, M Zipperer, Staufen im Breisgau, Germany) for 30 s on ice. Samples were then centrifuged 10 min at 12000 g at 4°C and supernatants were lyophilised and reconstituted in LiOH buffer (pH 2.2). The intracellular amino-acid content was determined by HPLC analysis with a Biochrom 20 amino acid analyser (Amersham Pharmacia Biotech, Little Chalfont, UK), using a high resolution column (Bio 20 Peek Lithium) and the physiological fluid chemical kit (Amersham Pharmacia Biotech) for elution. The column effluent was mixed with ninhydrin, passed through the high-temperature reaction coil and read by the photometer unit. Data were expressed as mmol l−1 serum or nmol mg−1 of tissue.
Glutamine synthetase activity
To assess liver GS activity, samples were homogenised in a solution containing 50 mM imidazole-HCl, pH 6.8, 0.5 mM EDTA, 1 mM DTT, supplemented with protease inhibitors (Complete, Mini, EDTA-free, Roche, Basel, Swizterland) for 30 s on ice. Samples were then sonicated on ice (Sonicator Ultrasonic Processor XL, Misonix, Farmingdale, NY, USA) and centrifuged for 30 min at 12000 g at 4°C. After quantification (Bio-Rad Protein Assay), protein concentration of the supernatant was adjusted to 1 mg ml−1. An aliquot of 150 μl was used for the assay in a solution of 50 mM imidazole-HCl (pH 6.8), 50 mM Gln, 25 mM hydroxylamine, 25 mM sodium arsenate, 2 mM MnCl2, and 0.16 mM ADP. After incubation at 37 °C for 30 min, the reaction was stopped by the addition of a solution containing 2.42% FeCl3 and 1.45% TCA in 1.82% HCl. Precipitates were removed by centrifugation (2000 r.p.m. for 5 min) and supernatants were read at 540 nm using a spectrophotometer (Helios-γ Spectronic, Thermo Electron Corporation, Cambridge, UK). Values of GS activity were expressed as pmol of γ-glutamylhydroxamate min−1 μg−1 protein.
Histopathology and immunostaining
The tumour samples were fixed in buffered formalin and embedded in paraffin. Section (4 μm) were stained with H&E for histological analysis. Immunohistochemical analyses were performed on all available specimens using the following primary antibodies: β-Catenin (CTNNB1, BD Transduction Laboratories, Franklin Lakes, NJ, USA; 1 : 1000), GS (BD Transduction Laboratories; 1 : 10000), Ki67 (mouse monoclonal, Dako Italia S.p.A., Cernusco sul Naviglio, Italy; 1 : 75), vimentin (clone V9, Roche, 1 : 1000), E-cadherin (clone 36, Roche, 1 : 1000), cytokeratin 7 (CK7, clone SP52, Roche, 1 : 1000), cytokeratin 19 (CK19, clone A53-B/A2.26, Roche, 1 : 1000), COX2 (clone SP21 Roche, 1 : 1000). After deparaffinisation and rehydratation, sections were treated with 3% hydrogen peroxide. For antigen retrieval, sections were treated with 10 mM citrate buffer at pH 6.0 in a microwave oven. Immunoreactions were developed with the HRP Polymer (Advance HRP Kit, Dako) and 3-31Diaminobenzidine. Adequate positive and negative controls were used in each set of reactions. Reactions were performed on an automated immunostainer BenchMark XT (Roche).
β-Catenin and Glutamine Synthetase reactivity was scored as previously described (Dal Bello et al, 2010). For the calculation of Ki67 index, slides were counterstained in hematoxylin (Mayer’s Hemalaun, AppliChem GmbH, Darmstadt, Germany) for 10 s and rinsed in running tap water for 10 min. Ki67 index was calculated counting a minimum of 10 randomly selected high-power ( × 400) fields containing representative sections of tumour and calculated as the percentage of positively stained cells to total cells (Burcombe et al, 2006).
Gene and protein expression
Gene expression was evaluated with RT-qPCR using 1 μg of total RNA extracted with the GenElute Mammalian mRNA Miniprep Kit (Sigma Aldrich Srl, Milan, Italy). PCR reactions were performed in a Rotor-Gene 3000 thermocycler (Corbett Research, Rotor-Gene 3000, version 5.0.60, Mortlake, Australia) using the GoTaq qPCR Master Mix (Promega Italia Srl, Milan, Italy) and 5 pmol of each primer. The primers used were 5′-GATTGCCTTCTGTTCAGTGTCT-3′ (for) and 5′-GGGTCAACTACCGCCAACC-3′ (rev), for Asparagine Synthetase (ASNS); 5′-TCATCTTGCATCGTGTGTGTG-3′ (for) and 5′-CTTCAGACCATTCTCCTCCGG-3′-(rev) for GS (GLUL); 5′-ATGAAGAAGGCCGAAATGGGA-3′ (for) and 5′-TGCTTGGTGGGGTAGGAGTAG-3′ (rev) for Solute carrier family 38 member A2 (SLC38A2); 5′-TGGTCTCCTGGATCATGTGG-3′ (for) and 5′-TTTGCGGGTGAAGAGGAAGT-3′ (rev) for Solute carrier family 1 member A5 (SLC1A5); 5′-GCAGCCATCAGGTAAGCCAAG-3′ (for) and 5′-AGCGGACCCTCAGAAGAA AGC-3′ (rev) for Ribosomal protein L15 (RPL15).
The reaction consisted of 35 cycles including a denaturation step at 95 °C for 30 s, followed by separate annealing (30 s, 55 °C–58 °C) and extension (30 s, 72°C) steps. Fluorescence was monitored at the end of each extension step. A no-template, no-reverse-transcriptase control was included in each experiment. At the end of the amplification cycles, a melting curve analysis was added. Data analysis was performed according to the Relative Standard Curve Method (Bustin 2000).
For the analysis of protein expression, cells were lysed in RIPA buffer containing a cocktail of protease inhibitors (Complete, Mini, EDTA-free, Roche). Lysates were transferred in tubes, sonicated for 5 s, centrifuged at 12 000 g for 10 min at 4 °C and aliquots of 35 μg of proteins were mixed with Laemmli buffer 4 × , warmed at 95 °C for 5 min and loaded on a 10% gel for SDS-PAGE. After electrophoresis, proteins were transferred to PVDF membranes (Immobilon-P, Millipore Spa, Milan, Italy). Non-specific binding sites were blocked with an incubation of 2 h at RT in 5% non-fat dried milk in TBS-Tween solution. The blots were then incubated at 4 °C overnight with the following antibodies diluted in a 5% BSA TBS-Tween solution: anti-GS (mouse, monoclonal, 1 : 1500, BD Transduction Laboratories), anti-β-catenin (mouse, monoclonal, 1 : 1000, Dako), anti p70S6K1 phosphoT389 (rabbit, monoclonal, 1 : 1000, Cell Signaling, Danvers, MA, USA), anti-ASNS (rabbit, polyclonal, 1 : 1000, Santa-Cruz Biotechnology, Santa Cruz, CA, USA), anti-vimentin (rabbit, polyclonal, 1 : 1000, Cell Signaling), anti-E-cadherin (rabbit, polyclonal, 1 : 1000, Cell Signaling), anti-SLUG (rabbit, polyclonal, 1 : 1000, Cell Signaling), anti-GADPH (rabbit, polyclonal, 1 : 4000, Sigma Aldrich Srl). After washing, blots were exposed for 1 h at room temperature to HRP-conjugated anti mouse or anti-rabbit antibody (Cell Signaling) diluted 1 : 10000 in blocking solution. Immunoreactivity was visualised with Immobilon Western Chemiluminescent HRP Substrate (Millipore Spa).
Immunofluorescence
Cells were seeded on 4-chamber Culture Slides (Falcon, Becton & Dickinson Company, San Jose, CA, USA) at a density of 4·104 cells cm−2. When 70% of confluence was reached, cells were rinsed twice in PBS and fixed for 10 min in 3.7% paraformaldehyde in PBS. After two further rinses, cells were permeabilised for 7 min in 0.1% Triton X-100 in PBS. Cells were then incubated for 1 h in blocking solution (5% of BSA, 10% of goat serum, and 0.3 M glycine in PBS) followed by an overnight incubation at 4 °C with anti-β-catenin monoclonal antibody (DakoCytomation Spa, Milan, Italy; M35359, 1 : 200 in PBS 5% BSA), two washes in PBS, and an additional 1-h incubation with an Alexa Fluor 488 anti-rabbit IgG antibody (Invitrogen, Paisley, UK, 1 : 400). After one rinse, cells were incubated for 30 min with a 100 μg ml−1 RNase A solution, and then with a 1 μg ml−1 solution of propidium iodide in PBS for 10 min to stain nuclei. Cells were observed with a confocal microscope Zeiss 510 LSM Meta (Carl Zeiss SpA, Arese, Milan, Italy), using an oil × 60 objective. λex of 488 nm and 540 nm and λem of 545 nm and 595 nm were used for Alexa Fluor 488 and propidium iodide, respectively.
Statistics
Data were expressed as means ±s.d. For statistical analysis, two-tail Student’s t-test for unpaired samples or one-way ANOVA were used. GraphPad Prism 5.0 (GraphPad Software Inc., La Jolla, CA, USA) was used for all the statistical analyses and P-values <0.05 were considered statistically significant.
Results
The combined treatment with Asparaginase and MSO depletes blood glutamine and asparagine
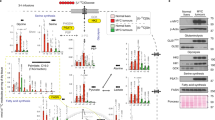
The combined treatment with 5 U g−1 CRIS and 10 mg kg−1 MSO (three times a week for 3 weeks) was well tolerated by athymic nude mice, with only a minor, although significant, decrease in body weight and no significant alterations in liver and spleen weights (Figure 1A). Treatment caused a marked decrease in the liver content of Gln and, more evidently, Asn (Figure 1B). In liver homogenates of the treated animals, MSO was detected (Figure 1B), demonstrating the tissue uptake of the inhibitor. The hepatic activity of GS (Figure 1C), was lowered by >97% compared with control. Serum ALT and albumin, as well as serum urea and creatinine (Supplementary Table S1), were not significantly different in control and treated animals (Figure 1D).
Biochemical effects of the combined treatment. Male nude mice were treated with CRIS and MSO for 3 weeks. Body, liver and spleen weights (A) were measured at the end of the treatment. Data are means ±s.d. (n=6) **P<0.01; NS, not significant, as assessed with one-way ANOVA. Liver amino acids (B), Glutamine synthetase activity (C) and serum albumin and ALT (D) in control and treated animals. Data represent means ±s.d. (n=3). *P<0.05; NS, not significant, as assessed with one-way ANOVA. ND, not detectable.
Glutaminolytic treatment suppresses the growth of HCC cell xenografts
The growth of HepG2 xenografts was significantly reduced after 19 days of treatments. Tumour size was reduced by 51% and 60% by single treatment with CRIS or MSO, respectively (Figure 2A), whereas it was completely suppressed by the combined treatment. At histopathological analysis no viable tumour cells were detectable in this group of animals and, instead, fibrous tissue with a lympho-macrophagic infiltrate was observed at the injection site consistent with complete tumour regression (Figure 3B, lower left). The tumour weight of treated animals was also reduced by 34% with CRIS and 61% with MSO (Figure 2B and C).
Effects of CRIS and MSO on HepG2 xenografts. Male athymic nude mice were subcutaneously engrafted with HepG2 cells. Treatments started 8 days after injection. (A) Tumour growth was monitored with caliper. (B) After 19 days of treatment, mice were killed and tumours were dissected and weighted. For (A and B), data represent means ±s.d. (n=5, n=4 for MSO group). *P<0.05, *** P<0.001, as assessed with a two-tail Student’s t-test for unpaired data. ND, not detectable.
Effects of CRIS and MSO on HepG2 xenografts. Male athymic nude mice were subcutaneously engrafted with HepG2 cells (same experiment of Figure 2). (A) Immunohistochemical detection of E-cadherin and β-catenin expression in HepG2 xenografts at the time of killing. Original magnification × 200. Bars, 50 μm. (B and C) Immunohistochemical detection of GS expression in xenografts (B) and the liver (C). Original magnification × 200. Bars, 50 μm. (D, E) Serum (D) and tumour (E) amino acids. Data represent means ±s.d. (n=4). *P<0.05, ***P<0.001 vs control, as assessed with a two-tail Student’s t-test for unpaired data. ND, not detectable.
The HepG2 xenografts (Figure 3A) showed a diffuse nuclear expression of β-catenin and a corresponding loss of membrane expression of E-cadherin. The pattern and the intensity of immunoreactivity were not modified by the different treatment regimens (not shown). HepG2 tumours were also intensely positive for GS (Figure 3B, top left), and the expression of the enzyme was not appreciably increased by either CRIS (Figure 3B, top right) or MSO (Figure 3B, bottom right). Glutamine synthetase-positive cells were not detected in the tissue surrounding the injection site of animals treated with CRIS and MSO, confirming that the grafted cells were completely cleared by the treatment (Figure 3B, bottom left). The liver tissue of control mice showed expression of GS in a single row of hepatocytes around the central hepatic venule. The number of perivenular hepatocytes stained with GS was increased in the animals receiving the combination of CRIS + MSO (Figure 3C). To assess the potential emergence of resistant tumour clones with features of progenitor liver cells and/or epithelial-mesenchymal transition, we performed on all available tumour samples immunostaining for markers of hepatocellular stemness (vimentin, CK7, CK19, (van Zijl et al, 2009) or enzymes known to be modulated by glutamine depletion in other tumour models (COX2, (Singh et al, 2012)). A variable expression of all these markers was observed irrespective of tumour size or treatment schedule (data not shown).
Serum amino acids in the four experimental conditions are shown in Figure 3D. Crisantaspase, alone or in combination with MSO, caused a substantial depletion of serum Asn and Gln. MSO alone did not affect serum Asn or Gln, but produced a significant decrease in serum Glu compared with untreated controls. Moreover, CRIS significantly decrease the content of Asn, Glu and Gln in the tumour tissue (Figure 3E).
The combined treatment hinders the growth of xenografts of CTNNB1-mutated, asparaginase-resistant HC-AFW1 cells
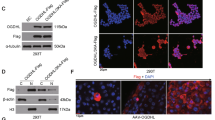
To ascertain if the behaviour exhibited by HepG2 cells was shared by other CTNNB1-mutated HCC cell lines, we investigated the sensitivity to Gln depletion of HC-AFW1 cells. These cells harbour a CTNNB1 deletion in exon 3, distinct from that present in HepG2 cells (Armeanu-Ebinger et al, 2012); however, in both cell lines the mutations result in the nuclear accumulation of β-catenin (Figure 4A). Although the growth of HepG2 cells was markedly hindered by CRIS (Tardito et al, 2011), HC-AFW1 cells were substantially unaffected (Figure 4B) and both lines were insensitive to MSO alone. On the contrary, in the presence of CRIS, MSO dramatically reduced the growth of both cell lines.
Effects of CRIS and MSO in HC-AFW1 cells and xenografts. (A) β-catenin in HC-AFW1 (left) and HepG2 cells (right). Green, β-catenin; Red, Propidium iodide. Bars, 20 μm. (B) Cell number was assessed after a 72-h incubation with CRIS (1 U ml−1) and/or MSO (1 mM). ***P<0.001 vs control (n=4). (C) Gene expression in HCC cells incubated in normal growth medium. mRNA content was expressed relatively to HepG2 cells. **P<0.01 HC-AFW1 vs HepG2 (n=4). A two-tail Student’s t-test for unpaired data was performed for data in (A and B). (D) Western blots of β-catenin, p70S6K1 (phosphoT389), ASNS and GS were performed on cells treated with CRIS (C) or CRIS + MSO (CM) – untreated cells. (E–H) NSG mice were engrafted with HC-AFW1 cells. (E) Tumour weights at the time of killing. (F) Dissected tumours. For each group, the tumour with the weight closest to the group mean is shown. (G) Relative tumour volume, calculated as the ratio between the volume after the last treatment and the volume before the first treatment. (H) Ki67 index. One-way ANOVA analysis was performed for data in (E, G and H).
To gain further insight on the differential effect of treatment on HepG2 and HC-AFW1 cells, the expression of genes involved in Gln metabolism and transport was investigated (Figure 4C). The GLUL gene, coding for GS, was more expressed in HC-AFW1 than in HepG2 cells, whereas ASNS, SLC38A2 (SNAT2 transporter) and SLC1A5 (ASCT2 transporter) were more expressed in HepG2 cells. The higher expression of GS in HC-AFW1 cells was also confirmed by western blot analysis (Figure 4D).
Glutamine synthetase expression was increased by treatment with CRIS or CRIS + MSO, and this effect was more evident in HepG2 than in HC-AFW1 cells. This is likely correlated with the depletion of intracellular Gln that leads to GS stabilisation (Tardito et al, 2011). Indeed, a 24-h treatment with CRIS lowered cell Gln from 75 nmoles mg−1 of protein, under control conditions, to undetectable levels (<1 nmole mg−1 of protein) in HepG2 cells, and from 51 nmoles mg−1 of protein to 16 nmoles mg−1 of protein in HC-AFW1 cells. The different degree of intracellular Gln depletion was also reflected by the different behaviour of the phosphorylated form of p70S6K1, a bona fide readout for mTOR activity, which was decreased by CRIS in HepG2 but not in HC-AFW1 cells (Figure 4D). Interestingly, as already described by our group (Tardito et al, 2012), in CRIS-treated cells, MSO causes a paradoxical activation of mTOR that is more evident in HepG2 than in HC-AFW1 cells.
When treatments were performed on HC-AFW1 xenografts (Panels E–H), only the combined treatment with CRIS + MSO produced a significant decrease (60%) in the mean weight of tumours at the time of killing (Figure 4E and F). Crisantaspase or, at a lesser degree, MSO alone also decreased tumour weight, without reaching statistical significance. However, when the tumour volumes were matched to their respective initial value, all treatments caused a significant delay in tumour growth (Figure 4G). The Ki67 index, an indicator of proliferative activity of the tumour, was significantly lowered by CRIS or CRIS + MSO, but not by MSO alone (Figure 4H). The expression of markers of epithelial-mesenchymal transition (vimentin, E-cadherin and SLUG) was not consistently changed by treatments also in HC-AFW1 tumours (Supplementary Figure S1).
Discussion
In this report, we show that two CTNNB1-mutated HCC models are sensitive to Gln depletion, obtained in vivo with the administration of CRIS, a drug in clinical use for Acute Lymphoblastic Leukaemia, and of the irreversible GS inhibitor MSO. The effect of CRIS treatment was differentially evident in the two cell lines, which harbour distinct CTNNB1 mutations, the HepG2 cells being more sensitive than the HC-AFW1 cells both in vitro and in vivo.
The combined treatment with CRIS and MSO exhibits only a mild toxicity in vivo as shown by a minor decrease in body weight in nude mice (Figure 1), confirmed in NSG mice (results not shown). Indices of liver, kidney and muscle function, such as serum albumin, ALT, urea and creatinine levels, are not significantly altered. However, given the possibility that glutamine depletion and inhibition of GS activity have detrimental effects on central nervous and immune systems, the toxicity of the treatment deserves further investigation in different models.
The hypothesised biochemical mechanisms for the antitumour effect of the treatment are shown in Figure 5. Besides asparagine hydrolysis, it is known that CRIS hydrolyses extracellular Gln ((Avramis 2012; Covini et al, 2012) and Figure 3D), and MSO inhibits GS activity, thus preventing intracellular synthesis of Gln. As a result, the cell content of Gln decreases at levels not permissive for cell proliferation, and HCC growth is hampered.
In animals with HepG2 xenografts treated with CRIS alone, tumour Gln is decreased by 50%. It is likely that this partial effect is due to the rescue of plasma Gln after the initial depletion that follows CRIS administration. Indeed, in leukaemia patients treated with bacterial asparaginases, only transient Gln depletion can be observed (Ollenschlager et al, 1988). Nonetheless, a significant delay in HepG2 and HC-AFW1 xenograft growth is observed in animals treated with CRIS alone, suggesting that even a partial Gln depletion has severe metabolic consequences for CTNNB1-mutated HCC cells.
Cancer cells acquire metabolic properties significantly different from those of normal cells, such as dysregulated glucose metabolism, stimulated fatty acid synthesis and enhanced glutaminolysis (DeBerardinis et al, 2007; Zhao et al, 2013). In many cancer models, these alterations are driven by oncogene activation or inactivating mutations of oncosuppressors. The β-catenin/Wnt pathway is activated in a significant proportion of HCC, but it has not been so far associated with major differences in the central carbon metabolism (Beyoglu et al, 2013), although the activation of this pathway is associated with distinctive transcriptional, pathological and clinical features (Boyault et al, 2007); yet, CTNNB1 mutations are strongly associated with the induction of genes involved in Gln metabolism (Cadoret et al, 2002), suggesting that the mutated CTNNB1 rewires cell metabolism towards a more Gln-dependent phenotype. Previous work from our (Tardito et al, 2011) as well as from other groups (Cheng et al, 2011) indicated that CTNBB1-mutated HepG2 cells are very sensitive to Gln starvation in vitro. The present report demonstrates that this sensitivity is also maintained in vivo. On the basis of these data, we have hypothesised that CTNNB1 mutations promote Gln addiction in HCC.
Another CTNNB1-mutated model, the HC-AFW1 cells (Figure 4A–D), differently from HepG2 cells, are resistant to CRIS in vitro and, consistently, maintain appreciable intracellular levels of Gln even when the extracellular amino acid is completely depleted. This behaviour is likely due to the higher expression of GS in HC-AFW1 cells. The divergent sensitivity to CRIS of HepG2 and HC-AFW1 cells suggests that different CTNNB1 mutations may have distinct effects on Gln metabolism in tumour cells. The low expression of transport systems for Gln, like SNAT2 and ASCT2 (Figure 4C), may also contribute to shield the intracellular compartment from Gln depletion, slowing the efflux of the amino acid from HC-AFW1 cells. The residual Gln content, maintained in this cell model, preserves the activity of mTOR, which is instead progressively lowered in CRIS-treated HepG2 cells (Figure 4D). HC-AFW1 cells, when xenografted in mice, are sensitive to the combined treatment with CRIS and MSO. Interestingly, when tumour volumes before killing (Figure 4G) were matched and normalised to the initial tumour volume size at the beginning of the treatments, CRIS and MSO, even when used alone, showed significant effects on tumour growth. Thus HC-AFW1 cells seem more sensitive to Gln depletion in vivo than in vitro. This paradox may be explained considering that the extracellular concentration of Gln is markedly higher in the culture medium (2 mM) than in serum (0.6 mM). Moreover, the HC-AFW1 cell line was derived from a paediatric HCC, a tumour that has distinctive epidemiology, aetiology, clinical behaviour and response to therapy compared with adult HCC (Emre and McKenna, 2004). Therefore, biological differences with the HepG2 cell line are to be expected despite the sharing of CTNNB1 mutations.
The possibility to hinder HCC growth through a metabolic approach based on amino-acid depletion has been recently investigated (Zhang et al, 2013), and it has been demonstrated that the knockdown of ASNS markedly increased the sensitivity to CRIS, both in vitro and in vivo. In our case, CRIS-resistant HC-AFW1 cells express much less ASNS than CRIS-sensitive HepG2 cells, indicating that CRIS resistance is not always linked to high ASNS expression and that additional factors determine CRIS antitumour effect. However, it should be noted that CRIS resistance, as evaluated in leukaemia cells, is a complex phenomenon (Aslanian and Kilberg, 2001), possibly attributable to the contribution of different mechanisms.
In summary, the results presented in this report indicate that the growth of human CTNNB1-mutated HCC xenografts is delayed by Gln restriction. Even the sole, likely transient depletion of circulating Gln, achieved by CRIS administration, is able to hinder tumour growth. Increasing evidence indicates that targeting tumour metabolism constitutes a promising strategy and may widen the therapeutic window of traditional chemotherapeutic agents (Zhao et al, 2013). Compared with pharmacological inhibitors of β-catenin or anti-β-catenin siRNA, which have been tested on HCC cells (Behari et al, 2007; Zeng et al, 2007), glutamine-depleting drugs act downstream of β-catenin on one of its targets, thus possibly causing fewer side effects on normal tissues that need β-catenin activation (Nejak-Bowen and Monga, 2011).
Further experiments will be needed to elucidate the relationships between β-catenin pathway and Gln addiction in HCC and, in particular, if the approach described in this contribution is also effective in HCC with wild type CTNNB1. However, the data presented suggest that Gln depletion may constitute a pharmacologically achievable, metabolic approach to control CTNNB1-mutated HCC.
Change history
09 September 2014
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Armeanu-Ebinger S, Wenz J, Seitz G, Leuschner I, Handgretinger R, Mau-Holzmann UA, Bonin M, Sipos B, Fuchs J, Warmann SW (2012) Characterisation of the cell line HC-AFW1 derived from a pediatric hepatocellular carcinoma. PLoS One 7: e38223.
Aslanian AM, Kilberg MS (2001) Multiple adaptive mechanisms affect asparagine synthetase substrate availability in asparaginase-resistant MOLT-4 human leukaemia cells. Biochem J 358: 59–67.
Avramis VI (2012) Asparaginases: biochemical pharmacology and modes of drug resistance. Anticancer Res 32: 2423–2437.
Behari J, Zeng G, Otruba W, Thompson MD, Muller P, Micsenyi A, Sekhon SS, Leoni L, Monga SP (2007) R-Etodolac decreases beta-catenin levels along with survival and proliferation of hepatoma cells. J Hepatol 46: 849–857.
Beyoglu D, Imbeaud S, Maurhofer O, Bioulac-Sage P, Zucman-Rossi J, Dufour JF, Idle JR (2013) Tissue metabolomics of hepatocellular carcinoma: tumor energy metabolism and the role of transcriptomic classification. Hepatology 58: 229–238.
Boyault S, Rickman DS, de Reynies A, Balabaud C, Rebouissou S, Jeannot E, Herault A, Saric J, Belghiti J, Franco D, Bioulac-Sage P, Laurent-Puig P, Zucman-Rossi J (2007) Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology 45: 42–52.
Burcombe R, Wilson GD, Dowsett M, Khan I, Richman PI, Daley F, Detre S, Makris A (2006) Evaluation of Ki-67 proliferation and apoptotic index before, during and after neoadjuvant chemotherapy for primary breast cancer. Breast Cancer Res 8: R31.
Bustin SA (2000) Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 25: 169–193.
Cadoret A, Ovejero C, Terris B, Souil E, Levy L, Lamers WH, Kitajewski J, Kahn A, Perret C (2002) New targets of beta-catenin signaling in the liver are involved in the glutamine metabolism. Oncogene 21: 8293–8301.
Cheng T, Sudderth J, Yang C, Mullen AR, Jin ES, Mates JM, DeBerardinis RJ (2011) Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proc Natl Acad Sci USA 108: 8674–8679.
Chiang DY, Villanueva A, Hoshida Y, Peix J, Newell P, Minguez B, LeBlanc AC, Donovan DJ, Thung SN, Sole M, Tovar V, Alsinet C, Ramos AH, Barretina J, Roayaie S, Schwartz M, Waxman S, Bruix J, Mazzaferro V, Ligon AH, Najfeld V, Friedman SL, Sellers WR, Meyerson M, Llovet JM (2008) Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res 68: 6779–6788.
Chiu M, Tardito S, Barilli A, Bianchi MG, Dall’Asta V, Bussolati O (2012) Glutamine stimulates mTORC1 independent of the cell content of essential amino acids. Amino Acids 43: 2561–2567.
Cieply B, Zeng G, Proverbs-Singh T, Geller DA, Monga SP (2009) Unique phenotype of hepatocellular cancers with exon-3 mutations in beta-catenin gene. Hepatology 49: 821–831.
Covini D, Tardito S, Bussolati O, Chiarelli LR, Pasquetto MV, Digilio R, Valentini G, Scotti C (2012) Expanding targets for a metabolic therapy of cancer: L-asparaginase. Recent Pat Anticancer Drug Discov 7: 4–13.
Dal Bello B, Rosa L, Campanini N, Tinelli C, Torello Viera F, D'Ambrosio G, Rossi S, Silini EM (2010) Glutamine synthetase immunostaining correlates with pathologic features of hepatocellular carcinoma and better survival after radiofrequency thermal ablation. Clin Cancer Res 16: 2157–2166.
DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB (2007) Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci USA 104: 19345–19350.
El-Serag HB (2011) Hepatocellular carcinoma. N Engl J Med 365: 1118–1127.
Emre S, McKenna GJ (2004) Liver tumors in children. Pediatr Transplant 8: 632–638.
Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM (2010) Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 127: 2893–2917.
Hoshida Y, Villanueva A, Kobayashi M, Peix J, Chiang DY, Camargo A, Gupta S, Moore J, Wrobel MJ, Lerner J, Reich M, Chan JA, Glickman JN, Ikeda K, Hashimoto M, Watanabe G, Daidone MG, Roayaie S, Schwartz M, Thung S, Salvesen HB, Gabriel S, Mazzaferro V, Bruix J, Friedman SL, Kumada H, Llovet JM, Golub TR (2008) Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med 359: 1995–2004.
Legoix P, Bluteau O, Bayer J, Perret C, Balabaud C, Belghiti J, Franco D, Thomas G, Laurent-Puig P, Zucman-Rossi J (1999) Beta-catenin mutations in hepatocellular carcinoma correlate with a low rate of loss of heterozygosity. Oncogene 18: 4044–4046.
Nejak-Bowen KN, Monga SP (2011) Beta-catenin signaling, liver regeneration and hepatocellular cancer: sorting the good from the bad. Semin Cancer Biol 21: 44–58.
Ollenschlager G, Roth E, Linkesch W, Jansen S, Simmel A, Modder B (1988) Asparaginase-induced derangements of glutamine metabolism: the pathogenetic basis for some drug-related side-effects. Eur J Clin Invest 18: 512–516.
Roessler S, Long EL, Budhu A, Chen Y, Zhao X, Ji J, Walker R, Jia HL, Ye QH, Qin LX, Tang ZY, He P, Hunter KW, Thorgeirsson SS, Meltzer PS, Wang XW (2012) Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology 142: 957–966 e912.
Rotoli BM, Uggeri J, Dall’Asta V, Visigalli R, Barilli A, Gatti R, Orlandini G, Gazzola GC, Bussolati O (2005) Inhibition of glutamine synthetase triggers apoptosis in asparaginase-resistant cells. Cell Physiol Biochem 15: 281–292.
Shariff MI, Cox IJ, Gomaa AI, Khan SA, Gedroyc W, Taylor-Robinson SD (2009) Hepatocellular carcinoma: current trends in worldwide epidemiology, risk factors, diagnosis and therapeutics. Expert Rev Gastroenterol Hepatol 3: 353–367.
Singh B, Tai K, Madan S, Raythatha MR, Cady AM, Braunlin M, Irving LR, Bajaj A, Lucci A (2012) Selection of metastatic breast cancer cells based on adaptability of their metabolic state. PLoS One 7: e36510.
Tardito S, Chiu M, Franchi-Gazzola R, Dall’Asta V, Comi P, Bussolati O (2012) The non-proteinogenic amino acids L-methionine sulfoximine and DL-phosphinothricin activate mTOR. Amino Acids 42: 2507–2512.
Tardito S, Chiu M, Uggeri J, Zerbini A, Da Ros F, Dall'Asta V, Missale G, Bussolati O (2011) L-Asparaginase and inhibitors of glutamine synthetase disclose glutamine addiction of beta-catenin-mutated human hepatocellular carcinoma cells. Curr Cancer Drug Targets 11: 929–943.
Tardito S, Uggeri J, Bozzetti C, Bianchi MG, Rotoli BM, Franchi-Gazzola R, Gazzola GC, Gatti R, Bussolati O (2007) The inhibition of glutamine synthetase sensitizes human sarcoma cells to L-asparaginase. Cancer Chemother Pharmacol 60: 751–758.
van Zijl F, Zulehner G, Petz M, Schneller D, Kornauth C, Hau M, Machat G, Grubinger M, Huber H, Mikulits W (2009) Epithelial-mesenchymal transition in hepatocellular carcinoma. Future Oncol 5: 1169–1179.
White BD, Chien AJ, Dawson DW (2012) Dysregulation of Wnt/beta-catenin signaling in gastrointestinal cancers. Gastroenterology 142: 219–232.
Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, Thompson CB (2008) Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci USA 105: 18782–18787.
Workman P, Aboagye EO, Balkwill F, Balmain A, Bruder G, Chaplin DJ, Double JA, Everitt J, Farningham DA, Glennie MJ, Kelland LR, Robinson V, Stratford IJ, Tozer GM, Watson S, Wedge SR, Eccles SA (2010) Guidelines for the welfare and use of animals in cancer research. Br J Cancer 102: 1555–1577.
Zeng G, Apte U, Cieply B, Singh S, Monga SP (2007) siRNA-mediated beta-catenin knockdown in human hepatoma cells results in decreased growth and survival. Neoplasia 9: 951–959.
Zhang B, Dong LW, Tan YX, Zhang J, Pan YF, Yang C, Li MH, Ding ZW, Liu LJ, Jiang TY, Yang JH, Wang HY (2013) Asparagine synthetase is an independent predictor of surgical survival and a potential therapeutic target in hepatocellular carcinoma. Br J Cancer 109: 14–23.
Zhao Y, Butler EB, Tan M (2013) Targeting cellular metabolism to improve cancer therapeutics. Cell Death Dis 4: e532.
Acknowledgements
This work has been funded by a grant of Associazione ONLUS Centro per la prevenzione e la terapia delle epatiti virali e delle loro complicanze, Parma, Italy, to OB. Crisantaspase was a generous gift of EUSA Pharma, an internal division of Jazz Pharmaceuticals, UK.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Chiu, M., Tardito, S., Pillozzi, S. et al. Glutamine depletion by crisantaspase hinders the growth of human hepatocellular carcinoma xenografts. Br J Cancer 111, 1159–1167 (2014). https://doi.org/10.1038/bjc.2014.425
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2014.425
Keywords
This article is cited by
-
Ammonia scavenger and glutamine synthetase inhibitors cocktail in targeting mTOR/β-catenin and MMP-14 for nitrogen homeostasis and liver cancer
Medical Oncology (2023)
-
Combinatorial antitumor effects of amino acids and epigenetic modulations in hepatocellular carcinoma cell lines
Naunyn-Schmiedeberg's Archives of Pharmacology (2021)
-
Hepatoblastoma: glutamine depletion hinders cell viability in the embryonal subtype but high GLUL expression is associated with better overall survival
Journal of Cancer Research and Clinical Oncology (2021)
-
Glutamine synthetase is necessary for sarcoma adaptation to glutamine deprivation and tumor growth
Oncogenesis (2019)
-
Metabolic rearrangements in primary liver cancers: cause and consequences
Nature Reviews Gastroenterology & Hepatology (2019)