Abstract
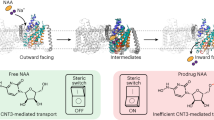
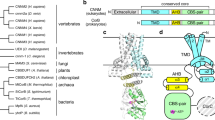
Nucleosides are required for DNA and RNA synthesis, and the nucleoside adenosine has a function in a variety of signalling processes1,2. Transport of nucleosides across cell membranes provides the major source of nucleosides in many cell types and is also responsible for the termination of adenosine signalling. As a result of their hydrophilic nature, nucleosides require a specialized class of integral membrane proteins, known as nucleoside transporters (NTs), for specific transport across cell membranes. In addition to nucleosides, NTs are important determinants for the transport of nucleoside-derived drugs across cell membranes3,4,5. A wide range of nucleoside-derived drugs, including anticancer drugs (such as Ara-C and gemcitabine) and antiviral drugs (such as zidovudine and ribavirin), have been shown to depend, at least in part, on NTs for transport across cell membranes4,6,7,8,9,10,11,12,13. Concentrative nucleoside transporters, members of the solute carrier transporter superfamily SLC28, use an ion gradient in the active transport of both nucleosides and nucleoside-derived drugs against their chemical gradients. The structural basis for selective ion-coupled nucleoside transport by concentrative nucleoside transporters is unknown. Here we present the crystal structure of a concentrative nucleoside transporter from Vibrio cholerae in complex with uridine at 2.4 Å. Our functional data show that, like its human orthologues, the transporter uses a sodium-ion gradient for nucleoside transport. The structure reveals the overall architecture of this class of transporter, unravels the molecular determinants for nucleoside and sodium binding, and provides a framework for understanding the mechanism of nucleoside and nucleoside drug transport across cell membranes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
King, A. E., Ackley, M. A., Cass, C. E., Young, J. D. & Baldwin, S. A. Nucleoside transporters: from scavengers to novel therapeutic targets. Trends Pharmacol. Sci. 27, 416–425 (2006)
Rose, J. B. & Coe, I. R. Physiology of nucleoside transporters: back to the future. Physiology (Bethesda) 23, 41–48 (2008)
Cano-Soldado, P. et al. Compensatory effects of the human nucleoside transporters on the response to nucleoside-derived drugs in breast cancer MCF7 cells. Biochem. Pharmacol. 75, 639–648 (2008)
Damaraju, V. L. et al. Nucleoside anticancer drugs: the role of nucleoside transporters in resistance to cancer chemotherapy. Oncogene 22, 7524–7536 (2003)
Jordheim, L. P. & Dumontet, C. Review of recent studies on resistance to cytotoxic deoxynucleoside analogues. Biochim. Biophys. Acta 1776, 138–159 (2007)
Errasti-Murugarren, E. & Pastor-Anglada, M. Drug transporter pharmacogenetics in nucleoside-based therapies. Pharmacogenomics 11, 809–841 (2010)
Mackey, J. R., Baldwin, S. A., Young, J. D. & Cass, C. E. Nucleoside transport and its significance for anticancer drug resistance. Drug Resist. Updat. 1, 310–324 (1998)
Mackey, J. R. et al. Immunohistochemical variation of human equilibrative nucleoside transporter 1 protein in primary breast cancers. Clin. Cancer Res. 8, 110–116 (2002)
Mackey, J. R. et al. Functional nucleoside transporters are required for gemcitabine influx and manifestation of toxicity in cancer cell lines. Cancer Res. 58, 4349–4357 (1998)
Nagai, K., Nagasawa, K. & Fujimoto, S. Uptake of the anthracycline pirarubicin into mouse M5076 ovarian sarcoma cells via a sodium-dependent nucleoside transport system. Cancer Chemother. Pharmacol. 55, 222–230 (2005)
Pastor-Anglada, M., Felipe, A. & Casado, F. J. Transport and mode of action of nucleoside derivatives used in chemical and antiviral therapies. Trends Pharmacol. Sci. 19, 424–430 (1998)
Pastor-Anglada, M. et al. Nucleoside transporters in chronic lymphocytic leukaemia. Leukemia 18, 385–393 (2004)
Zhang, J. et al. The role of nucleoside transporters in cancer chemotherapy with nucleoside drugs. Cancer Metastasis Rev. 26, 85–110 (2007)
Gray, J. H., Owen, R. P. & Giacomini, K. M. The concentrative nucleoside transporter family, SLC28. Pflugers Arch. 447, 728–734 (2004)
Molina-Arcas, M., Casado, F. J. & Pastor-Anglada, M. Nucleoside transporter proteins. Curr. Vasc. Pharmacol. 7, 426–434 (2009)
Ritzel, M. W. et al. Molecular identification and characterization of novel human and mouse concentrative Na+-nucleoside cotransporter proteins (hCNT3 and mCNT3) broadly selective for purine and pyrimidine nucleosides (system cib). J. Biol. Chem. 276, 2914–2927 (2001)
Loewen, S. K. et al. Functional characterization of a H+/nucleoside co-transporter (CaCNT) from Candida albicans, a fungal member of the concentrative nucleoside transporter (CNT) family of membrane proteins. Yeast 20, 661–675 (2003)
Loewen, S. K. et al. Transport of physiological nucleosides and anti-viral and anti-neoplastic nucleoside drugs by recombinant Escherichia coli nucleoside-H+ cotransporter (NupC) produced in Xenopus laevis oocytes. Mol. Membr. Biol. 21, 1–10 (2004)
Xiao, G., Wang, J., Tangen, T. & Giacomini, K. M. A novel proton-dependent nucleoside transporter, CeCNT3, from Caenorhabditis elegans. Mol. Pharmacol. 59, 339–348 (2001)
Hamilton, S. R. et al. Subcellular distribution and membrane topology of the mammalian concentrative Na+-nucleoside cotransporter rCNT1. J. Biol. Chem. 276, 27981–27988 (2001)
Slugoski, M. D. et al. Substituted cysteine accessibility method analysis of human concentrative nucleoside transporter hCNT3 reveals a novel discontinuous region of functional importance within the CNT family motif (G/A)XKX3NEFVA(Y/M/F). J. Biol. Chem. 284, 17281–17292 (2009)
Zhang, J. et al. Cysteine-accessibility analysis of transmembrane domains 11–13 of human concentrative nucleoside transporter 3. Biochem. J. 394, 389–398 (2006)
Cao, Y. et al. Crystal structure of a phosphorylation-coupled saccharide transporter. Nature 473, 50–54 (2011)
Yernool, D., Boudker, O., Jin, Y. & Gouaux, E. Structure of a glutamate transporter homologue from Pyrococcus horikoshii. Nature 431, 811–818 (2004)
Loewen, S. K. et al. Identification of amino acid residues responsible for the pyrimidine and purine nucleoside specificities of human concentrative Na+ nucleoside cotransporters hCNT1 and hCNT2. J. Biol. Chem. 274, 24475–24484 (1999)
Slugoski, M. D. et al. Conserved glutamate residues Glu-343 and Glu-519 provide mechanistic insights into cation/nucleoside cotransport by human concentrative nucleoside transporter hCNT3. J. Biol. Chem. 284, 17266–17280 (2009)
Yao, S. Y. et al. Conserved glutamate residues are critically involved in Na+/nucleoside cotransport by human concentrative nucleoside transporter 1 (hCNT1). J. Biol. Chem. 282, 30607–30617 (2007)
Nayal, M. & Di Cera, E. Valence screening of water in protein crystals reveals potential Na+ binding sites. J. Mol. Biol. 256, 228–234 (1996)
Yamashita, A., Singh, S. K., Kawate, T., Jin, Y. & Gouaux, E. Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature 437, 215–223 (2005)
Krishnamurthy, H., Piscitelli, C. L. & Gouaux, E. Unlocking the molecular secrets of sodium-coupled transporters. Nature 459, 347–355 (2009)
Koszelak-Rosenblum, M. et al. Determination and application of empirically derived detergent phase boundaries to effectively crystallize membrane proteins. Protein Sci. 18, 1828–1839 (2009)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Sheldrick, G. M. A short history of SHELX. Acta Crystallogr. A 64, 112–122 (2008)
Terwilliger, T. C. & Berendzen, J. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 (1999)
Terwilliger, T. C. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 56, 965–972 (2000)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010)
Lee, S. Y., Letts, J. A. & MacKinnon, R. Functional reconstitution of purified human Hv1 H+ channels. J. Mol. Biol. 387, 1055–1060 (2009)
Acknowledgements
Data for this study were collected at beamlines SER-CAT BM22/ID22 and NE-CAT ID 24-C at the Advanced Photon Source. We thank R. MacKinnon and J. Butterwick for critical reading; R. Lefkowitz and A. Shukla for providing access and technical support for the radioactive flux assay; S. Lockless for advice on experiments; and C. Pemble for help with remote data collection. This work was supported by start-up funds from the Duke University Medical Center, the McKnight Endowment Fund for Neuroscience, the Alfred P. Sloan Foundation, the Klingenstein Fund, the Mallinckrodt foundation, the Basil O’Connor Starter Scholar Research Award 5-FY10-473 from the March of Dimes Foundation, and the National Institutes of Health Director’s New Innovator Award 1 DP2 OD008380-01 (all to S.-Y.L.).
Author information
Authors and Affiliations
Contributions
Z.J. expressed, purified and crystallized vcCNT. Z.J. performed radioactive flux and crosslinking experiments. C.-G.C. participated in part of the vcCNT crystallization and generated mutants for crystallization and functional studies. Z.J. and S.-Y.L. collected and processed the data, solved the structure, and wrote the paper. S.-Y.L. designed the study. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Table 1, Supplementary Figures 1-7, a Supplementary Discussion and additional references. (PDF 7882 kb)
Rights and permissions
About this article
Cite this article
Johnson, Z., Cheong, CG. & Lee, SY. Crystal structure of a concentrative nucleoside transporter from Vibrio cholerae at 2.4 Å. Nature 483, 489–493 (2012). https://doi.org/10.1038/nature10882
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10882
This article is cited by
-
Magnetic bacterial cellulose nanofibers for nucleoside recognition
Cellulose (2020)
-
Structures of human ENT1 in complex with adenosine reuptake inhibitors
Nature Structural & Molecular Biology (2019)
-
Calcium is an essential cofactor for metal efflux by the ferroportin transporter family
Nature Communications (2018)
-
Rosetta Broker for membrane protein structure prediction: concentrative nucleoside transporter 3 and corticotropin-releasing factor receptor 1 test cases
BMC Structural Biology (2017)
-
Structural insights into the elevator-like mechanism of the sodium/citrate symporter CitS
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.