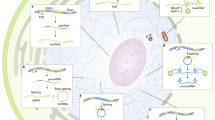
Key Points
-
We now understand that genomes in all three domains of life produce functional RNAs that do not encode proteins (non-coding (nc)RNAs), but do exert important influences on diverse cellular processes.
-
Most ncRNAs function with essential partner proteins (that is, as non-coding ribonucleoproteins; ncRNPs) and use cognate antisense elements to interact with target molecules. The small nuclear (sn)RNAs and small nucleolar (sno)RNAs are founding members of the family of ncRNAs that helped to establish these common paradigms.
-
Recent studies of the snRNPs and snoRNPs have revealed unexpectedly elaborate biogenesis pathways that will probably also be travelled by other ncRNPs.
-
snRNAs and snoRNAs can be transcribed from independent promoters (similar to mRNAs) or can be encoded within intronic sequences.
-
RNA function can require multiple partner proteins with roles that might include modulating RNA structure or securing an enzyme, as well as catalysing the reaction.
-
Assembling a functional complex seems to involve a series of non-functional intermediate states that are matured by a series of cellular factors along a defined physical pathway in the cell. These transport and assembly steps might serve as control points for the regulation of the activity of a given ncRNP.
Abstract
Recent advances have fuelled rapid growth in our appreciation of the tremendous number, diversity and biological importance of non-coding (nc)RNAs. Because ncRNAs typically function as ribonucleoprotein (RNP) complexes and not as naked RNAs, understanding their biogenesis is crucial to comprehending their regulation and function. The small nuclear and small nucleolar RNPs are two well studied classes of ncRNPs with elaborate assembly and trafficking pathways that provide paradigms for understanding the biogenesis of other ncRNPs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Cheng, J. et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308, 1149–1154 (2005).
Mattick, J. S. & Makunin, I. V. Non-coding RNA. Hum. Mol. Genet. 15, R17–R29 (2006).
Huttenhofer, A., Schattner, P. & Polacek, N. Non-coding RNAs: hope or hype? Trends Genet. 21, 289–297 (2005).
Storz, G., Altuvia, S. & Wassarman, K. M. An abundance of RNA regulators. Annu. Rev. Biochem. 74, 199–217 (2005).
Szymanski, M., Barciszewska, M. Z., Zywicki, M. & Barciszewski, J. Noncoding RNA transcripts. J. Appl. Genet. 44, 1–19 (2003).
Eddy, S. R. Computational genomics of noncoding RNA genes. Cell 109, 137–140 (2002).
Goodrich, J. A. & Kugel, J. F. Non-coding-RNA regulators of RNA polymerase II transcription. Nature Rev. Mol. Cell Biol. 7, 612–616 (2006).
Huttenhofer, A. & Schattner, P. The principles of guiding by RNA: chimeric RNA–protein enzymes. Nature Rev. Genet. 7, 475–482 (2006).
Valadkhan, S. snRNAs as the catalysts of pre-mRNA splicing. Curr. Opin. Chem. Biol. 9, 603–608 (2005).
Will, C. L. & Luhrmann, R. in The RNA World 3rd edn (eds Gesteland, R. F., Cech, T. R. & Atkins, A. J.) 369–400 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, 2006).
Nilsen, T. W. The spliceosome: the most complex macromolecular machine in the cell? Bioessays 25, 1147–1149 (2003).
Hernandez, N. Small nuclear RNA genes: a model system to study fundamental mechanisms of transcription. J. Biol. Chem. 276, 26733–26736 (2001).
Hernandez, N. & Weiner, A. M. Formation of the 3′ end of U1 snRNA requires compatible snRNA promoter elements. Cell 47, 249–258 (1986).
de Vegvar, H. E., Lund, E. & Dahlberg, J. E. 3′ end formation of U1 snRNA precursors is coupled to transcription from snRNA promoters. Cell 47, 259–266 (1986).
Baillat, D. et al. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 123, 265–276 (2005). These authors purified a long-sought complex required for 3′-end processing of pre-snRNA transcripts.
Dominski, Z., Yang, X. C. & Marzluff, W. F. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell 123, 37–48 (2005).
Kolev, N. G. & Steitz, J. A. Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes Dev. 19, 2583–2592 (2005). Together with reference 16, showed that core factors involved in mRNA polyadenylation form distinct complexes that are required for proper histone 3′-end maturation.
Steinmetz, E. J., Conrad, N. K., Brow, D. A. & Corden, J. L. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature 413, 327–331 (2001). The first paper to recognize the role of Nrd1 in preventing read-through transcription of non-polyadenylated snRNA and snoRNA genes in yeast.
Carroll, K. L., Pradhan, D. A., Granek, J. A., Clarke, N. D. & Corden, J. L. Identification of cis elements directing termination of yeast nonpolyadenylated snoRNA transcripts. Mol. Cell. Biol. 24, 6241–6252 (2004).
Steinmetz, E. J., Ng, S. B., Cloute, J. P. & Brow, D. A. cis- and trans-acting determinants of transcription termination by yeast RNA polymerase II. Mol. Cell. Biol. 26, 2688–2696 (2006).
Sheldon, K. E., Mauger, D. M. & Arndt, K. M. A requirement for the Saccharomyces cerevisiae Paf1 complex in snoRNA 3′ end formation. Mol. Cell 20, 225–236 (2005).
Thiebaut, M., Kisseleva-Romanova, E., Rougemaille, M., Boulay, J. & Libri, D. Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the Nrd1–Nab3 pathway in genome surveillance. Mol. Cell 23, 853–864 (2006).
Arigo, J. T., Eyler, D. E., Carroll, K. L. & Corden, J. L. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol. Cell 23, 841–851 (2006).
Schramm, L. & Hernandez, N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 16, 2593–2620 (2002).
Ohno, M., Segref, A., Bachi, A., Wilm, M. & Mattaj, I. W. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell 101, 187–198 (2000).
Segref, A., Mattaj, I. W. & Ohno, M. The evolutionarily conserved region of the U snRNA export mediator PHAX is a novel RNA-binding domain that is essential for U snRNA export. RNA 7, 351–360 (2001).
Ohno, M., Segref, A., Kuersten, S. & Mattaj, I. W. Identity elements used in export of mRNAs. Mol. Cell 9, 659–671 (2002).
Masuyama, K., Taniguchi, I., Kataoka, N. & Ohno, M. RNA length defines RNA export pathway. Genes Dev. 18, 2074–2085 (2004).
Terns, M. P. & Dahlberg, J. E. Retention and 5′ cap trimethylation of U3 snRNA in the nucleus. Science 264, 959–961 (1994).
Boulon, S. et al. PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol. Cell 16, 777–787 (2004).
Watkins, N. J. et al. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol. Cell 16, 789–798 (2004).
Terns, M. P., Grimm, C., Lund, E. & Dahlberg, J. E. A common maturation pathway for small nucleolar RNAs. EMBO J. 14, 4860–4871 (1995).
Speckmann, W., Narayanan, A., Terns, R. & Terns, M. P. Nuclear retention elements of U3 small nucleolar RNA. Mol. Cell. Biol. 19, 8412–8421 (1999).
Smith, K. P. & Lawrence, J. B. Interactions of U2 gene loci and their nuclear transcripts with Cajal (coiled) bodies: evidence for PreU2 within Cajal bodies. Mol. Biol. Cell 11, 2987–2998 (2000).
Matera, A. G. & Shpargel, K. B. Pumping RNA: nuclear bodybuilding along the RNP pipeline. Curr. Opin. Cell Biol. 18, 317–324 (2006).
Terns, M. P. & Terns, R. M. Macromolecular complexes: SMN — the master assembler. Curr. Biol. 11, R862–R864 (2001).
Eggert, C., Chari, A., Laggerbauer, B. & Fischer, U. Spinal muscular atrophy: the RNP connection. Trends Mol. Med. 12, 113–121 (2006).
Golembe, T. J., Yong, J. & Dreyfuss, G. Specific sequence features, recognized by the SMN complex, identify snRNAs and determine their fate as snRNPs. Mol. Cell. Biol. 25, 10989–11004 (2005).
Battle, D. J. et al. The Gemin5 protein of the SMN complex identifies snRNAs. Mol. Cell 23, 273–279 (2006).
Pillai, R. S. et al. Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev. 17, 2321–2333 (2003). Showed that LSM11 replaces SmD2 in the U7 core and demonstrated the existence of a distinct pool of SMN complexes that lack the canonical SmD1 and SmD2, but that contain LSM10 and LSM11.
Mouaikel, J., Verheggen, C., Bertrand, E., Tazi, J. & Bordonne, R. Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol. Cell 9, 891–901 (2002). Identified the 5′-cap hypermethylase Tgs1, the activity of which was later shown to be essential in metazoans.
Huang, Q. & Pederson, T. A human U2 RNA mutant stalled in 3′ end processing is impaired in nuclear import. Nucleic Acids Res. 27, 1025–1031 (1999).
Palacios, I., Hetzer, M., Adam, S. A. & Mattaj, I. W. Nuclear import of U snRNPs requires importin β. EMBO J. 16, 6783–6792 (1997).
Huber, J. et al. Snurportin1, an m3G-cap-specific nuclear import receptor with a novel domain structure. EMBO J. 17, 4114–4126 (1998).
Narayanan, U., Achsel, T., Luhrmann, R. & Matera, A. G. Coupled in vitro import of U snRNPs and SMN, the spinal muscular atrophy protein. Mol. Cell 16, 223–234 (2004). Showed that the SMN complex participates in nuclear import as an adaptor between the Sm core and importin-β.
Huber, J., Dickmanns, A. & Luhrmann, R. The importin-β binding domain of snurportin1 is responsible for the Ran- and energy-independent nuclear import of spliceosomal U snRNPs in vitro. J. Cell Biol. 156, 467–479 (2002).
Ospina, J. K. et al. Cross-talk between snurportin1 subdomains. Mol. Biol. Cell 16, 4660–4671 (2005).
Sleeman, J. E. & Lamond, A. I. Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr. Biol. 9, 1065–1074 (1999).
Stanek, D., Rader, S. D., Klingauf, M. & Neugebauer, K. M. Targeting of U4/U6 small nuclear RNP assembly factor SART3/p110 to Cajal bodies. J. Cell Biol. 160, 505–516 (2003).
Nesic, D., Tanackovic, G. & Kramer, A. A role for Cajal bodies in the final steps of U2 snRNP biogenesis. J. Cell Sci. 117, 4423–4433 (2004).
Schaffert, N., Hossbach, M., Heintzmann, R., Achsel, T. & Luhrmann, R. RNAi knockdown of hPrp31 leads to an accumulation of U4/U6 di-snRNPs in Cajal bodies. EMBO J. 23, 3000–3009 (2004).
Stanek, D. & Neugebauer, K. M. Detection of snRNP assembly intermediates in Cajal bodies by fluorescence resonance energy transfer. J. Cell Biol. 166, 1015–1025 (2004).
Tanackovic, G. & Kramer, A. Human splicing factor SF3a, but not SF1, is essential for pre-mRNA splicing in vivo. Mol. Biol. Cell 16, 1366–1377 (2005).
Brown, J. W. et al. Plant snoRNA database. Nucleic Acids Res. 31, 432–435 (2003).
Lestrade, L. & Weber, M. J. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 34, D158–D162 (2006).
Kiss, T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109, 145–148 (2002).
Henras, A. K., Dez, C. & Henry, Y. RNA structure and function in C/D and H/ACA s(no)RNPs. Curr. Opin. Struct. Biol. 14, 335–343 (2004).
Meier, U. T. The many facets of H/ACA ribonucleoproteins. Chromosoma 114, 1–14 (2005).
Terns, M. P. & Terns, R. M. Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin. Gene Expr. 10, 17–39 (2002).
Yu, Y. T., Terns, R. M. & Terns, M. P. in Fine-tuning of RNA Functions by Modification and Editing Vol. 12 (ed. Grosjean, H.) 223–262 (Topics in Current Genetics, New York, 2005).
Decatur, W. A. & Fournier, M. J. RNA-guided nucleotide modification of ribosomal and other RNAs. J. Biol. Chem. 278, 695–698 (2003).
Darzacq, X. et al. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J. 21, 2746–2756 (2002). Identified scaRNAs as modification-guide RNAs that localize in Cajal bodies.
Dennis, P. P., Omer, A. & Lowe, T. A guided tour: small RNA function in Archaea. Mol. Microbiol. 40, 509–519 (2001).
Uliel, S., Liang, X. H., Unger, R. & Michaeli, S. Small nucleolar RNAs that guide modification in trypanosomatids: repertoire, targets, genome organisation, and unique functions. Int. J. Parasitol. 34, 445–454 (2004).
Cavaille, J. et al. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl Acad. Sci. USA 97, 14311–14316 (2000).
Vitali, P. et al. ADAR2-mediated editing of RNA substrates in the nucleolus is inhibited by C/D small nucleolar RNAs. J. Cell Biol. 169, 745–753 (2005).
Kishore, S. & Stamm, S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science 311, 230–232 (2006).
Collins, K. The biogenesis and regulation of telomerase holoenzymes. Nature Rev. Mol. Cell Biol. 7, 484–494 (2006).
Huttenhofer, A. et al. RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J. 20, 2943–2953 (2001).
Kiss, A. M., Jady, B. E., Bertrand, E. & Kiss, T. Human box H/ACA pseudouridylation guide RNA machinery. Mol. Cell. Biol. 24, 5797–5807 (2004).
Omer, A. D., Ziesche, S., Ebhardt, H. & Dennis, P. P. In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc. Natl Acad. Sci. USA 99, 5289–5294 (2002). Established the essential components of the C/D RNP and its hierarchical assembly pathway.
Bortolin, M. L., Bachellerie, J. P. & Clouet-d'Orval, B. In vitro RNP assembly and methylation guide activity of an unusual box C/D RNA, cis-acting archaeal pre-tRNA(Trp). Nucleic Acids Res. 31, 6524–6535 (2003).
Rashid, R. et al. Functional requirement for symmetric assembly of archaeal box C/D small ribonucleoprotein particles. J. Mol. Biol. 333, 295–306 (2003).
Tran, E. J., Zhang, X. & Maxwell, E. S. Efficient RNA 2′-O-methylation requires juxtaposed and symmetrically assembled archaeal box C/D and C′/D′ RNPs. EMBO J. 22, 3930–3940 (2003).
Baker, D. L. et al. RNA-guided RNA modification: functional organization of the archaeal H/ACA RNP. Genes Dev. 19, 1238–1248 (2005).
Charpentier, B., Muller, S. & Branlant, C. Reconstitution of archaeal H/ACA small ribonucleoprotein complexes active in pseudouridylation. Nucleic Acids Res. 33, 3133–3144 (2005). References 75 and 76 defined the essential components and organization of the H/ACA RNP.
Wang, H., Boisvert, D., Kim, K. K., Kim, R. & Kim, S. H. Crystal structure of a fibrillarin homologue from Methanococcus jannaschii, a hyperthermophile, at 1.6 Å resolution. EMBO J. 19, 317–323 (2000).
Deng, L. et al. Structure determination of fibrillarin from the hyperthermophilic archaeon Pyrococcus furiosus. Biochem. Biophys. Res. Commun. 315, 726–732 (2004).
Aittaleb, M. et al. Structure and function of archaeal box C/D sRNP core proteins. Nature Struct. Biol. 10, 256–263 (2003). Provided insight into the structure and organization of a key subcomplex of the C/D RNP.
Suryadi, J., Tran, E. J., Maxwell, E. S. & Brown, B. A. 2nd. The crystal structure of the Methanocaldococcus jannaschii multifunctional L7Ae RNA-binding protein reveals an induced-fit interaction with the box C/D RNAs. Biochemistry 44, 9657–9672 (2005).
Moore, T., Zhang, Y., Fenley, M. O. & Li, H. Molecular basis of box C/D RNA-protein interactions; cocrystal structure of archaeal L7Ae and a box C/D RNA. Structure 12, 807–818 (2004).
Rashid, R. et al. Crystal structure of a Cbf5–Nop10–Gar1 complex and implications in RNA-guided pseudouridylation and dyskeratosis congenita. Mol. Cell 21, 249–260 (2006).
Manival, X. et al. Crystal structure determination and site-directed mutagenesis of the Pyrococcus abyssi aCBF5–aNOP10 complex reveal crucial roles of the C-terminal domains of both proteins in H/ACA sRNP activity. Nucleic Acids Res. 34, 826–839 (2006).
Hamma, T., Reichow, S. L., Varani, G. & Ferre-D'Amare, A. R. The Cbf5–Nop10 complex is a molecular bracket that organizes box H/ACA RNPs. Nature Struct. Mol. Biol. 12, 1101–1107 (2005).
Hamma, T. & Ferre-D'Amare, A. R. Structure of protein L7Ae bound to a K-turn derived from an archaeal box H/ACA sRNA at 1.8 Å resolution. Structure 12, 893–903 (2004).
Li, L. & Ye, K. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature 443, 302–307 (2006). References 82 and 86 describe the crystal structures of H/ACA RNP complexes and provide insight into the molecular basis of dyskeratosis congenita.
Mitchell, J. R., Wood, E. & Collins, K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402, 551–555 (1999).
Mochizuki, Y., He, J., Kulkarni, S., Bessler, M. & Mason, P. J. Mouse dyskerin mutations affect accumulation of telomerase RNA and small nucleolar RNA, telomerase activity, and ribosomal RNA processing. Proc. Natl Acad. Sci. USA 101, 10756–10761 (2004).
Marrone, A., Walne, A. & Dokal, I. Dyskeratosis congenita: telomerase, telomeres and anticipation. Curr. Opin. Genet. Dev. 15, 249–257 (2005).
Watkins, N. J. et al. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA 4, 1549–1568 (1998).
Wang, C., Query, C. C. & Meier, U. T. Immunopurified small nucleolar ribonucleoprotein particles pseudouridylate rRNA independently of their association with phosphorylated Nopp140. Mol. Cell. Biol. 22, 8457–8466 (2002).
Henras, A. K., Capeyrou, R., Henry, Y. & Caizergues-Ferrer, M. Cbf5p, the putative pseudouridine synthase of H/ACA-type snoRNPs, can form a complex with Gar1p and Nop10p in absence of Nhp2p and box H/ACA snoRNAs. RNA 10, 1704–1712 (2004).
Schultz, A., Nottrott, S., Watkins, N. J. & Luhrmann, R. Protein–protein and protein–RNA contacts both contribute to the 15.5K-mediated assembly of the U4/U6 snRNP and the box C/D snoRNPs. Mol. Cell. Biol. 26, 5146–5154 (2006).
Szewczak, L. B., DeGregorio, S. J., Strobel, S. A. & Steitz, J. A. Exclusive interaction of the 15.5 kD protein with the terminal box C/D motif of a methylation guide snoRNP. Chem. Biol. 9, 1095–1107 (2002).
Darzacq, X. et al. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J. Cell Biol. 173, 207–218 (2006).
Ballarino, M., Morlando, M., Pagano, F., Fatica, A. & Bozzoni, I. The cotranscriptional assembly of snoRNPs controls the biosynthesis of H/ACA snoRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 25, 5396–5403 (2005).
Yang, P. K. et al. Cotranscriptional recruitment of the pseudouridylsynthetase Cbf5p and of the RNA binding protein Naf1p during H/ACA snoRNP assembly. Mol. Cell. Biol. 25, 3295–3304 (2005). References 95 and 97 provided detailed evidence for the current model of co-transcriptional assembly of inactive pre-H/ACA RNPs, including the role of exchange factors.
Richard, P., Kiss, A. M., Darzacq, X. & Kiss, T. Cotranscriptional recognition of human intronic box H/ACA snoRNAs occurs in a splicing-independent manner. Mol. Cell. Biol. 26, 2540–2549 (2006).
Fatica, A., Dlakic, M. & Tollervey, D. Naf1 p is a box H/ACA snoRNP assembly factor. RNA 8, 1502–1514 (2002).
Yang, P. K., Rotondo, G., Porras, T., Legrain, P. & Chanfreau, G. The Shq1p–Naf1p complex is required for box H/ACA small nucleolar ribonucleoprotein particle biogenesis. J. Biol. Chem. 277, 45235–45242 (2002).
Dez, C., Noaillac-Depeyre, J., Caizergues-Ferrer, M. & Henry, Y. Naf1p, an essential nucleoplasmic factor specifically required for accumulation of box H/ACA small nucleolar RNPs. Mol. Cell. Biol. 22, 7053–7065 (2002).
Hoareau-Aveilla, C., Bonoli, M., Caizergues-Ferrer, M. & Henry, Y. hNaf1 is required for accumulation of human box H/ACA snoRNPs, scaRNPs, and telomerase. RNA 12, 832–840 (2006).
Morlando, M. et al. Coupling between snoRNP assembly and 3′ processing controls box C/D snoRNA biosynthesis in yeast. EMBO J. 23, 2392–2401 (2004).
Hirose, T. & Steitz, J. A. Position within the host intron is critical for efficient processing of box C/D snoRNAs in mammalian cells. Proc. Natl Acad. Sci. USA 98, 12914–12919 (2001).
Verheggen, C. et al. Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J. 21, 2736–2745 (2002).
Hirose, T., Shu, M. D. & Steitz, J. A. Splicing-dependent and-independent modes of assembly for intron-encoded box C/D snoRNPs in mammalian cells. Mol. Cell 12, 113–123 (2003).
Hirose, T. et al. A spliceosomal intron binding protein, IBP160, links position-dependent assembly of intron-encoded box C/D snoRNP to pre-mRNA splicing. Mol. Cell 23, 673–684 (2006).
Peng, W. T. et al. A panoramic view of yeast noncoding RNA processing. Cell 113, 919–933 (2003).
Hiley, S. L., Babak, T. & Hughes, T. R. Global analysis of yeast RNA processing identifies new targets of RNase III and uncovers a link between tRNA 5′ end processing and tRNA splicing. Nucleic Acids Res. 33, 3048–3056 (2005).
Houseley, J., LaCava, J. & Tollervey, D. RNA-quality control by the exosome. Nature Rev. Mol. Cell Biol. 7, 529–539 (2006).
LaCava, J. et al. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121, 713–724 (2005).
Vasiljeva, L. & Buratowski, S. Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol. Cell 21, 239–248 (2006).
King, T. H., Decatur, W. A., Bertrand, E., Maxwell, E. S. & Fournier, M. J. A well-connected and conserved nucleoplasmic helicase is required for production of box C/D and H/ACA snoRNAs and localization of snoRNP proteins. Mol. Cell. Biol. 21, 7731–7746 (2001).
Samarsky, D. A., Fournier, M. J., Singer, R. H. & Bertrand, E. The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J. 17, 3747–3757 (1998).
Richard, P. et al. A common sequence motif determines the Cajal body-specific localization of box H/ACA scaRNAs. EMBO J. 22, 4283–4293 (2003).
Narayanan, A., Speckmann, W., Terns, R. & Terns, M. P. Role of the box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli. Mol. Biol. Cell 10, 2131–2147 (1999).
Gall, J. G. The centennial of the Cajal body. Nature Rev. Mol. Cell Biol. 4, 975–980 (2003).
Cioce, M. & Lamond, A. I. Cajal bodies: a long history of discovery. Annu. Rev. Cell. Dev. Biol. 21, 105–131 (2005).
Gubitz, A. K., Feng, W. & Dreyfuss, G. The SMN complex. Exp. Cell Res. 296, 51–56 (2004).
Whitehead, S. E. et al. Determinants of the interaction of the spinal muscular atrophy disease protein SMN with the dimethylarginine-modified box H/ACA small nucleolar ribonucleoprotein GAR1. J. Biol. Chem. 277, 48087–48093 (2002).
Pellizzoni, L., Baccon, J., Charroux, B. & Dreyfuss, G. The survival of motor neurons (SMN) protein interacts with the snoRNP proteins fibrillarin and GAR1. Curr. Biol. 11, 1079–1088 (2001).
Jones, K. W. et al. Direct interaction of the spinal muscular atrophy disease protein SMN with the small nucleolar RNA-associated protein fibrillarin. J. Biol. Chem. 276, 38645–38651 (2001).
Narayanan, A. et al. Nucleolar localization signals of box H/ACA small nucleolar RNAs. EMBO J. 18, 5120–5130 (1999).
Lange, T. S., Ezrokhi, M., Amaldi, F. & Gerbi, S. A. Box H and box ACA are nucleolar localization elements of U17 small nucleolar RNA. Mol. Biol. Cell 10, 3877–3890 (1999).
Jady, B. E., Bertrand, E. & Kiss, T. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J. Cell Biol. 164, 647–652 (2004).
Fu, D. & Collins, K. Human telomerase and Cajal body ribonucleoproteins share a unique specificity of Sm protein association. Genes Dev. 20, 531–536 (2006).
Tomlinson, R. L., Ziegler, T. D., Supakorndej, T., Terns, R. M. & Terns, M. P. Cell cycle-regulated trafficking of human telomerase to telomeres. Mol. Biol. Cell 17, 955–965 (2006).
Jady, B. E., Richard, P., Bertrand, E. & Kiss, T. Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres. Mol. Biol. Cell 17, 944–954 (2006). References 127 and 128 revealed that trafficking of the human telomerase RNP is regulated as a function of the cell cycle.
Seto, A. G., Zaug, A. J., Sobel, S. G., Wolin, S. L. & Cech, T. R. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature 401, 177–180 (1999).
Li, C. F. et al. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell 126, 93–106 (2006).
Pontes, O. et al. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell 126, 79–92 (2006).
Briese, M., Esmaeili, B. & Sattelle, D. B. Is spinal muscular atrophy the result of defects in motor neuron processes? Bioessays 27, 946–957 (2005).
Wattendorf, D. J. & Muenke, M. Prader–Willi syndrome. Am. Fam. Physician 72, 827–830 (2005).
Cahill, N. M. et al. Site-specific cross-linking analyses reveal an asymmetric protein distribution for a box C/D snoRNP. EMBO J. 21, 3816–3828 (2002).
Filipowicz, W. & Pogacic, V. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 14, 319–327 (2002).
Eliassen, K. A., Baldwin, A., Sikorski, E. M. & Hurt, M. M. Role for a YY1-binding element in replication-dependent mouse histone gene expression. Mol. Cell. Biol. 18, 7106–7118 (1998).
Acknowledgements
We apologize to our colleagues whose work has not been cited or discussed in full, owing to space constraints. We thank current and past members of the Matera and Terns groups who have contributed to the work in our laboratories. M.P.T. and R.M.T. are grateful to C. Glover for continued scientific discourse. This work was supported by National Institutes of Health (National Institute of Neurological Disorders and Stroke and National Institute of General Medical Sciences) and Muscular Dystrophy Association grants to A.G.M. and by grants from the National Institutes of Health (National Institute of General Medical Sciences and National Cancer Institute) and the Nora L. Redman Fund to M.P.T. and R.M.T.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Non-coding (nc)RNA
-
A functional RNA molecule that does not code for a protein (that is, it is not an mRNA).
- Adaptor protein
-
Bridges proteins that link specific cargoes to their cognate transport receptors.
- Export/import receptor
-
Collectively called karyopherins, these proteins facilitate transport across nuclear pores. Import factors are called importins and export factors are called exportins. Receptors primarily mediate interactions with nuclear pores but can also bind cargo directly.
- Cajal bodies
-
Intranuclear structures that function as ribonucleoprotein-assembly, trafficking and remodelling centres.
- Perichromatin fibril
-
(PF). A fine structure, which is visible only under the electron microscope, that is located adjacent to transcriptionally active chromatin and is thought to be a site of active pre-mRNA processing.
- Interchromatin granule cluster
-
(IGC).Corresponds to a nuclear domain visible as a speckle under the light microscope. IGCs function as storage sites for splicing factors that can be recruited to perichromatin fibrils.
- Peptidyl transferase centre
-
Catalytic centre of the ribosome that forms peptide bonds during protein translation.
- mRNA-decoding centre
-
Region of the ribosome involved in the selection of transfer RNAs that correspond to mRNA codons during protein translation.
- Kink-turn (k-turn) motif
-
A common RNA structural motif that is bound by a family of related proteins, including L7Ae, resulting in a sharp bend (or kink) in the RNA helix.
- TRAMP complex
-
A nuclear polyadenylation complex consisting of Trf4 (a poly(A) polymerase), Air2 (a zinc-knuckle protein) and Mtr4 (an RNA helicase). TRAMP functions together with the exosome as a quality-control mechanism to stimulate the degradation of various aberrant target RNAs.
- Exosome
-
A complex of 3′→5′ exonucleases that has important roles in RNA processing and turnover.
Rights and permissions
About this article
Cite this article
Matera, A., Terns, R. & Terns, M. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol 8, 209–220 (2007). https://doi.org/10.1038/nrm2124
Issue Date:
DOI: https://doi.org/10.1038/nrm2124
This article is cited by
-
Intronic small nucleolar RNAs regulate host gene splicing through base pairing with their adjacent intronic sequences
Genome Biology (2023)
-
Extranodal lymphoma: pathogenesis, diagnosis and treatment
Molecular Biomedicine (2023)
-
The forgotten variable? Does the euthanasia method and sample storage condition influence an organisms transcriptome – a gene expression analysis on multiple tissues in pigs
BMC Genomics (2023)
-
Developmental programming: adverse sexually dimorphic transcriptional programming of gestational testosterone excess in cardiac left ventricle of fetal sheep
Scientific Reports (2023)
-
Non-coding RNAs in human health and disease: potential function as biomarkers and therapeutic targets
Functional & Integrative Genomics (2023)