Abstract
Cultured cell lines are useful models in biomedical research that characterize metabolic responses to various stimuli (e.g., pathogens, toxins, or drugs/chemicals) and explore the underlying mechanisms. However, data from cell metabolomic studies must be normalized to the amount of cells, which is dependent on diverse treatments. The currently used methods of cell counting and protein assay involve extra work and delay the quenching of intracellular metabolism. To develop a convenient, alternative approach, in this study, intracellular metabolites were extracted from a series amount of cultured adherent cells and profiled by gas chromatography–time-of-flight mass spectrometry (GC–TOFMS). The GC–TOFMS signal intensities for 11 intracellular markers present in two different cell lines showed good linearity with the protein content, with inositol and pantothenate most promising (correlation coefficient > 0.970). Despite the various amounts of cells, the data normalized to the metabolic markers and protein amounts showed similar effectiveness, resulted in better separation of the two cell lines, closer clustering within each group(cell line) on a principal components analysis scores plot, and had lower relative standard deviations for intracellular metabolites than those of the non-normalized data, suggesting that these markers were effective indicators of cell amounts and independent of cell lines.

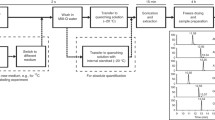
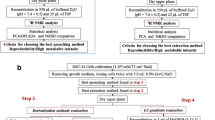
A schematic strategy for identifying intracellular metabolic markers for use as cell amount indicators in data normalization




Similar content being viewed by others
References
Mamas M, Dunn WB, Neyses L, Goodacre R (2011) The role of metabolites and metabolomics in clinically applicable biomarkers of disease. Arch Toxicol 85:5–17. doi:10.1007/s00204-010-0609-6
Cuperlovic-Culf M, Barnett DA, Culf AS, Chute I (2010) Cell culture metabolomics: applications and future directions. Drug Discov Today 15(15–16):610–621
Khoo Soo Hean G, Al-Rubeai M (2007) Metabolomics as a complementary tool in cell culture. Biotechnol Appl Biochem 47(2):71
Varghese RS, Cheema A, Cheema P, Bourbeau M, Tuli L, Zhou B, Jung M, Dritschilo A, Ressom HW (2010) Analysis of LC-MS data for characterizing the metabolic changes in response to radiation. J Proteome Res 9(5):2786–2793
Ritter JB, Wahl AS, Freund S, Genzel Y, Reichl U (2010) Metabolic effects of influenza virus infection in cultured animal cells: intra- and extracellular metabolite profiling. BMC Syst Biol 4:61
Jozefczuk S, Klie S, Catchpole G, Szymanski J, Cuadros-Inostroza A, Steinhauser D, Selbig J, Willmitzer L (2010) Metabolomic and transcriptomic stress response of Escherichia coli. Mol Syst Biol 6:364
Bayet-Robert M, Morvan D, Chollet P, Barthomeuf C (2010) Pharmacometabolomics of docetaxel-treated human MCF7 breast cancer cells provides evidence of varying cellular responses at high and low doses. Breast Cancer Res Treat 120(3):613–626
Klawitter J, Shokati T, Moll V, Christians U (2010) Effects of lovastatin on breast cancer cells: a proteo-metabonomic study. Breast Cancer Res 12(2):R16
Tiziani S, Lodi A, Khanim FL, Viant MR, Bunce CM, Günther UL, Cordes N (2009) Metabolomic profiling of drug responses in acute myeloid leukaemia cell lines. PLoS ONE 4(1):e4251
Ramanathan A, Schreiber SL (2009) Direct control of mitochondrial function by mTOR. Proc Natl Acad Sci USA 106(52):22229–22232
Heinemann M, Zenobi R (2011) Single cell metabolomics. Curr Opin Biotechnol 22:26–31. doi:10.1016/j.copbio.2010.09.008
Dettmer K, Nurnberger N, Kaspar H, Gruber MA, Almstetter MF, Oefner PJ (2011) Metabolite extraction from adherently growing mammalian cells for metabolomics studies: optimization of harvesting and extraction protocols. Anal Bioanal Chem 399:1127–1139. doi:10.1007/s00216-010-4425-x
Duarte IF, Marques J, Ladeirinha AF, Rocha C, Lamego I, Calheiros R, Silva TM, Marques MP, Melo JB, Carreira IM, Gil AM (2009) Analytical approaches toward successful human cell metabolome studies by NMR spectroscopy. Anal Chem 81(12):5023–5032
Teng Q, Huang W, Collette TW, Ekman DR, Tan C (2008) A direct cell quenching method for cell-culture based metabolomics. Metabolomics 5(2):199–208
Sellick CA, Hansen R, Maqsood AR, Dunn WB, Stephens GM, Goodacre R, Dickson AJ (2009) Effective quenching processes for physiologically valid metabolite profiling of suspension cultured mammalian cells. Anal Chem 81(1):174–183
Shin MH, Lee do Y, Liu KH, Fiehn O, Kim KH (2010) Evaluation of sampling and extraction methodologies for the global metabolic profiling of Saccharophagus degradans. Anal Chem 82(15):6660–6666
Dietmair S, Timmins NE, Gray PP, Nielsen LK, Kromer JO (2010) Towards quantitative metabolomics of mammalian cells: development of a metabolite extraction protocol. Anal Biochem 404(2):155–164
Ritter JB, Genzel Y, Reichl U (2008) Simultaneous extraction of several metabolites of energy metabolism and related substances in mammalian cells: optimization using experimental design. Anal Biochem 373(2):349–369
Danielsson AP, Moritz T, Mulder H, Spegel P (2010) Development and optimization of a metabolomic method for analysis of adherent cell cultures. Anal Biochem 404(1):30–39
Teahan O, Bevan CL, Waxman J, Keun HC (2010) Metabolic signatures of malignant progression in prostate epithelial cells. Int J Biochem Cell Biol. doi:10.1016/j.biocel.2010.07.003
Lyer VV, Ovacik MA, Androulakis IP, Roth CM, Ierapetritou MG (2010) Transcriptional and metabolic flux profiling of triadimefon effects on cultured hepatocytes. Toxicol Appl Pharmacol 248(3):165–177
Jiye A, Trygg J, Gullberg J, Johansson AI, Jonsson P, Antti H, Marklund SL, Moritz T (2005) Extraction and GC/MS analysis of the human blood plasma metabolome. Anal Chem 77(24):8086–8094
Jonsson P, Johansson AI, Gullberg J, Trygg J, Jiye A, Grung B, Marklund S, Sjostrom M, Antti H, Moritz T (2005) High-throughput data analysis for detecting and identifying differences between samples in GC/MS-based metabolomic analyses. Anal Chem 77(17):5635–5642
Eker C, Rydell R, Svanberg K, Andersson-Engels S (2001) Multivariate analysis of laryngeal fluorescence spectra recorded in vivo. Lasers Surg Med 28(3):259–266
Trygg J, Holmes E, Lundstedt T (2007) Chemometrics in metabonomics. J Proteome Res 6(2):469–479
Horio M, Yamauchi A, Moriyama T, Imai E, Orita Y (1997) Osmotic regulation of amino acids and system A transport in Madin-Darby canine kidney cells. Am J Physiol 272(3 Pt 1):C804–C809
Acknowledgments
This work is supported by grants from the National Key New Drug Creation Special Programs (2009ZX09304-001 and 2009ZX09502-004), National Natural Science Foundation of the People’s Republic of China (81072692, 30630076, 40821140541, and 30870086), the National 11th 5-Year Technology Supporting Program of the People’s Republic of China (No. 2006BAI08B04), the Jiangsu Nature Science Fund (BK2008038), and the National “973” Key Fundamental programs (2011CB505300 and 2011CB505303).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 1332 kb)
Rights and permissions
About this article
Cite this article
Cao, B., Aa, J., Wang, G. et al. GC–TOFMS analysis of metabolites in adherent MDCK cells and a novel strategy for identifying intracellular metabolic markers for use as cell amount indicators in data normalization. Anal Bioanal Chem 400, 2983–2993 (2011). https://doi.org/10.1007/s00216-011-4981-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-011-4981-8