Abstract
Background
Citalopram and escitalopram, selective serotonin reuptake inhibitors, are primarily metabolized by cytochrome P450 (CYP) 2C19, which is a highly polymorphic enzyme known to cause inter-individual differences in pharmacokinetics. However, the impact of CYP2C19 polymorphisms on citalopram or escitalopram exposure has yet to be fully clarified, especially with regard to the quantitative impact of the CYP2C19*17 allele.
Objective
The objective of this study was to quantify the effect of functional CYP2C19 allele variants on citalopram/escitalopram exposure.
Methods
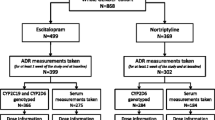
We performed a systematic review and meta-analysis with a structured search algorithm and eligibility criteria for including related studies, calculating the change of citalopram or escitalopram exposure associated with CYP2C19*2, *3, and *17 as compared with CYP2C19*1 using fixed-effect and random-effects models. Assessment of publication bias was performed by means of funnel plots and sensitivity analysis using meta-regressions. The pre-defined review protocol was registered at the PROSPERO international prospective register of systematic reviews, registration number CRD42013004106.
Results
Sixteen studies from 14 publications met the inclusion criteria. Eligible studies included 847 patients from psychiatric patient trials and 140 healthy subjects from pharmacokinetic studies. Compared to subjects with the EM/EM (CYP2C19*1/*1) genotype, the exposure to (es)citalopram increased by 95 % (95 % CI 40–149, p < 0.0001) in the poor metabolizer (PM)/PM (CYP2C19*2 or *3/*2 or *3), 30 % (95 % CI 4–55, p < 0.05) in the extensive metabolizer (EM)/PM (CYP2C19*1/*2 or *3), and 25 % (95 % CI 1–49, p < 0.05) in the ultrarapid metabolizer (UM)/PM (CYP2C19*17/*2 or *3) groups. In contrast, the exposure to (es)citalopram decreased by 36 % (95 % CI 27–46, p < 0.0001) in the UM/UM (CYP2C19*17/*17) and by 14 % (95 % CI 1–27, p < 0.05) in the UM/EM (CYP2C19*17/*1).
Interpretation
This is the first meta-analysis based on a systematic review of accumulated information that addresses the relationship between CYP2C19 genotypes and the exposure to citalopram or escitalopram. All functional CYP2C19 genotype groups demonstrated significant effects on (es)citalopram exposure. The findings based on our pooled analysis are likely to help in understanding the inter-individual variability in the exposure to citalopram and escitalopram in psychiatric patients and to facilitate dose selection, particularly for the homozygous carriers of CYP2C19*2 or *3 (loss of function) and CYP2C19*17 (gain of function) alleles. The results could improve individualization of citalopram or escitalopram therapy and could also be used for physiologically based pharmacokinetic modeling as well as pharmacokinetic/pharmacodynamic modeling.




Similar content being viewed by others
References
Brøsen K, Naranjo CA. Review of pharmacokinetic and pharmacodynamic interaction studies with citalopram. Eur Neuropsychopharmacol. 2001;11(4):275–83.
Owens MJ, Knight DL, Nemeroff CB. Second-generation SSRIs: human monoamine transporter binding profile of escitalopram and R-fluoxetine. Biol Psychiatry. 2001;50(5):345–50.
Drewes P, Tjijssen I, Mengel H, Larsen F. A single-dose cross-over pharmacokinetic study comparing racemic citalopram (40 mg) with the S-enantiomer of citalopram (escitalopram, 20 mg) in healthy male subjects. 41st Annual Meeting of the New Clinical Drug Evaluation Unit; 28–31 May 2001; Phoenix.
Burke WJ. Escitalopram. Expert Opin Investig Drugs. 2002;11(10):1477–86.
Rochat B, Amey M, Gillet M, Meyer UA, Baumann P. Identification of three cytochrome P450 isozymes involved in N-demethylation of citalopram enantiomers in human liver microsomes. Pharmacogenetics. 1997;7(1):1–10.
Chang M, Tybring G, Dahl ML, Götharson E, Sagar M, Seensalu R, et al. Interphenotype differences in disposition and effect on gastrin levels of omeprazole—suitability of omeprazole as a probe for CYP2C19. Br J Clin Pharmacol. 1995;39(5):511–8.
Stingl JC, Brockmöller J, Viviani R. Genetic variability of drug-metabolizing enzymes: the dual impact on psychiatric therapy and regulation of brain function. Mol Psychiatry. 2013;18(3):273–87.
Sim SC, Risinger C, Dahl ML, Aklillu E, Christensen M, Bertilsson L, et al. A common novel CYP2C19 gene variant causes ultrarapid drug metabolism relevant for the drug response to proton pump inhibitors and antidepressants. Clin Pharmacol Ther. 2006;79(1):103–13.
Ohlsson Rosenborg S, Mwinyi J, Andersson M, Baldwin RM, Pedersen RS, Sim SC, et al. Kinetics of omeprazole and escitalopram in relation to the CYP2C19*17 allele in healthy subjects. Eur J Clin Pharmacol. 2008;64(12):1175–9.
Rudberg I, Mohebi B, Hermann M, Refsum H, Molden E. Impact of the ultrarapid CYP2C19*17 allele on serum concentration of escitalopram in psychiatric patients. Clin Pharmacol Ther. 2008;83(2):322–7.
Tsai MH, Lin KM, Hsiao MC, Shen WW, Lu ML, Tang HS, et al. Genetic polymorphisms of cytochrome P450 enzymes influence metabolism of the antidepressant escitalopram and treatment response. Pharmacogenomics. 2010;11(4):537–46.
de Vos A, van der Weide J, Loovers HM. Association between CYP2C19*17 and metabolism of amitriptyline, citalopram and clomipramine in Dutch hospitalized patients. Pharmacogenomics J. 2011;11(5):359–67.
Huezo-Diaz P, Perroud N, Spencer EP, Smith R, Sim S, Virding S, et al. CYP2C19 genotype predicts steady state escitalopram concentration in GENDEP. J Psychopharmacol. 2012;26(3):398–407.
Thase ME, Haight BR, Richard N, Rockett CB, Mitton M, Modell JG, et al. Remission rates following antidepressant therapy with bupropion or selective serotonin reuptake inhibitors: a meta-analysis of original data from 7 randomized controlled trials. J Clin Psychiatry. 2005;66(8):974–81.
Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, Rennie D, et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis of Observational Studies in Epidemiology (MOOSE) group. JAMA. 2000;283(15):2008–12.
Lindh JD, Holm L, Andersson ML, Rane A. Influence of CYP2C9 genotype on warfarin dose requirements–a systematic review and meta-analysis. Eur J Clin Pharmacol. 2009;65(4):365–75.
R Development Core Team. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2012. ISBN 3-900051-07-0, http://www.R-project.org/. Accessed 14 Aug 2014
Rudberg I, Hendset M, Uthus LH, Molden E, Refsum H. Heterozygous mutation in CYP2C19 significantly increases the concentration/dose ratio of racemic citalopram and escitalopram (S-citalopram). Ther Drug Monit. 2006;28(1):102–5.
Herrlin K, Yasui-Furukori N, Tybring G, Widén J, Gustafsson LL, Bertilsson L. Metabolism of citalopram enantiomers in CYP2C19/CYP2D6 phenotyped panels of healthy Swedes. Br J Clin Pharmacol. 2003;56(4):415–21.
Sindrup SH, Brøsen K, Hansen MG, Aaes-Jørgensen T, Overø KF, Gram LF. Pharmacokinetics of citalopram in relation to the sparteine and the mephenytoin oxidation polymorphisms. Ther Drug Monit. 1993;15(1):11–7.
Baumann P, Nil R, Souche A, Montaldi S, Baettig D, Lambert S, et al. A double-blind, placebo-controlled study of citalopram with and without lithium in the treatment of therapy-resistant depressive patients: a clinical, pharmacokinetic, and pharmacogenetic investigation. J Clin Psychopharmacol. 1996;16(4):307–14.
Carlsson B, Olsson G, Reis M, Walinder J, Nordin C, Lundmark J, et al. Enantioselective analysis of citalopram and metabolites in adolescents. Ther Drug Monit. 2001;23(6):658–64.
Yu BN, Chen GL, He N, Ouyang DS, Chen XP, Liu ZQ, et al. Pharmacokinetics of citalopram in relation to genetic polymorphism of CYP2C19. Drug Metab Dispos. 2003;31(10):1255–9.
Noehr-Jensen L, Zwisler ST, Larsen F, Sindrup SH, Damkier P, Nielsen F, et al. Impact of CYP2C19 phenotypes on escitalopram metabolism and an evaluation of pupillometry as a serotonergic biomarker. Eur J Clin Pharmacol. 2009;65(9):887–94.
Fudio S, Borobia AM, Piñana E, Ramírez E, Tabarés B, Guerra P, et al. Evaluation of the influence of sex and CYP2C19 and CYP2D6 polymorphisms in the disposition of citalopram. Eur J Pharmacol. 2010;626(2–3):200–4.
Jiang T, Rong Z, Xu Y, Chen B, Xie Y, Chen C, et al. Pharmacokinetics and bioavailability comparison of generic and branded citalopram 20 mg tablets: an open-label, randomized-sequence, two-period crossover study in healthy Chinese CYP2C19 extensive metabolizers. Clin Drug Investig. 2013;33(1):1–9.
Chang M, Dahl ML, Tybring G, Götharson E, Bertilsson L. Use of omeprazole as a probe drug for CYP2C19 phenotype in Swedish Caucasians: comparison with S-mephenytoin hydroxylation phenotype and CYP2C19 genotype. Pharmacogenetics. 1995;5(6):358–63.
Sim SC, Kacevska M, Ingelman-Sundberg M. Pharmacogenomics of drug-metabolizing enzymes: a recent update on clinical implications and endogenous effects. Pharmacogenomics J. 2013;13(1):1–11.
Acknowledgments
The study was financially supported by the Swedish Research Council (521-2012-2592). JDL holds a post-doctoral position financed by Karolinska Institutet and Stockholm County Council (KI/SLL).
Author contributions
Conceived and designed the experiments: JDL, MC, MLD. Performed the experiments: MC, GT, JDL. Analyzed the data: JDL, MC. Contributed reagents/materials/analysis tools: JDL, MC. Wrote the paper: MC, GT, MLD, JDL.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supporting Information
Protocol S1 Pre-registered study protocol. Meta-analysis study protocol pre-registered at the PROSPERO international register of systematic reviews (registration number CRD 42013003530). (PDF). A full list of extracted data items is presented in S2.
40262_2014_162_MOESM1_ESM.pdf
Supplemental Fig. 1. Funnel plots of the association between the estimated mean difference (MD) and its standard error in individual studies. The vertical line represents overall MD estimate, the center of plot derived by using fixed-effects model in absence of bias. The other two dotted lines represent 95 % confidence limits for expected distribution of studies in absence of heterogeneity between studies. Supplementary material 1 (PDF 59 kb)
40262_2014_162_MOESM2_ESM.pptx
Supplemental Fig. 2 Random effects meta-regressions. The dotted lines represent 95 % confidence intervals of regression lines (solid lines). Sizes of dots are proportional to the weight of each study in the regression model. Supplementary material 2 (PPTX 108 kb)
Rights and permissions
About this article
Cite this article
Chang, M., Tybring, G., Dahl, ML. et al. Impact of Cytochrome P450 2C19 Polymorphisms on Citalopram/Escitalopram Exposure: A Systematic Review and Meta-Analysis. Clin Pharmacokinet 53, 801–811 (2014). https://doi.org/10.1007/s40262-014-0162-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40262-014-0162-1