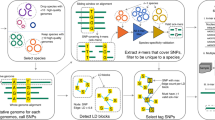
Abstract
The human body is colonized by a vast array of microbes, which form communities of bacteria, viruses and microbial eukaryotes that are specific to each anatomical environment. Every community must be studied as a whole because many organisms have never been cultured independently, and this poses formidable challenges. The advent of next-generation DNA sequencing has allowed more sophisticated analysis and sampling of these complex systems by culture-independent methods. These methods are revealing differences in community structure between anatomical sites, between individuals, and between healthy and diseased states, and are transforming our view of human biology.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Backhed, F., Ley, R. E., Sonnenburg, J. L., Peterson, D. A. & Gordon, J. I. Host–bacterial mutualism in the human intestine. Science 307, 1915–1920 (2005).
Foxman, B., Goldberg, D., Murdock, C., Xi, C. & Gilsdorf, J. R. Conceptualizing human microbiota: from multicelled organ to ecological community. Interdiscip. Perspect. Infect. Dis. 2008, 613979 (2008).
Possemiers, S., Bolca, S., Verstraete, W. & Heyerick, A. The intestinal microbiome: a separate organ inside the body with the metabolic potential to influence the bioactivity of botanicals. Fitoterapia 82, 53–66 (2011).
Shanahan, F. The host–microbe interface within the gut. Best Pract. Res. Clin. Gastroenterol. 16, 915–931 (2002).
Bruls, T. & Weissenbach, J. The human metagenome: our other genome? Hum. Mol. Genet. 20, R142–R148 (2011).
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010). This paper presents initial findings on the gut microbiome from the MetaHIT project.
Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012). This paper presents analysis of data from the HMP.
Stein, J. L., Marsh, T. L., Wu, K. Y., Shizuya, H. & DeLong, E. F. Characterization of uncultivated prokaryotes: isolation and analysis of a 40-kilobase-pair genome fragment from a planktonic marine archaeon. J. Bacteriol. 178, 591–599 (1996).
Vergin, K. L. et al. Screening of a fosmid library of marine environmental genomic DNA fragments reveals four clones related to members of the order Planctomycetales. Appl. Environ. Microbiol. 64, 3075–3078 (1998).
Olsen, G. J., Lane, D. J., Giovannoni, S. J., Pace, N. R. & Stahl, D. A. Microbial ecology and evolution: a ribosomal RNA approach. Annu. Rev. Microbiol. 40, 337–365 (1986).
Manichanh, C. et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut 55, 205–211 (2006).
Fredricks, D. N., Fiedler, T. L. & Marrazzo, J. M. Molecular identification of bacteria associated with bacterial vaginosis. N. Engl. J. Med. 353, 1899–1911 (2005).
Flint, H. J., Bayer, E. A., Rincon, M. T., Lamed, R. & White, B. A. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nature Rev. Microbiol. 6, 121–131 (2008).
Srikanth, C. V. & McCormick, B. A. Interactions of the intestinal epithelium with the pathogen and the indigenous microbiota: a three-way crosstalk. Interdiscip. Perspect. Infect. Dis. 2008, 626827 (2008).
Jakobsson, H. E. et al. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS ONE 5, e9836 (2010).
Dethlefsen, L., Huse, S., Sogin, M. L. & Relman, D. A. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 6, e280 (2008).
Miller, C. P., Bohnhoff, M. & Rifkind, D. The effect of an antibiotic on the susceptibility of the mouse's intestinal tract to Salmonella infection. Trans. Am. Clin. Climatol. Assoc. 68, 51–55 (1956).
Sekirov, I. et al. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect. Immun. 76, 4726–4736 (2008).
Croswell, A., Amir, E., Teggatz, P., Barman, M. & Salzman, N. H. Prolonged impact of antibiotics on intestinal microbial ecology and susceptibility to enteric Salmonella infection. Infect. Immun. 77, 2741–2753 (2009).
Mulligan, M. E. Epidemiology of Clostridium difficile-induced intestinal disease. Clin. Infect. Dis. 6, S222–S228 (1984).
Jarchum, I. & Pamer, E. G. Regulation of innate and adaptive immunity by the commensal microbiota. Curr. Opin. Immunol. 23, 353–360 (2011).
Marsland, B. J. Regulation of inflammatory responses by the commensal microbiota. Thorax 67, 93–94 (2012).
Wang, Z. et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63 (2011).
Gough, E., Shaikh, H. & Manges, A. R. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin. Infect. Dis. 53, 994–1002 (2011).
Brandt, L. J. & Reddy, S. S. Fecal microbiota transplantation for recurrent clostridium difficile infection. J. Clin. Gastroenterol. 45, S159–S167 (2011).
D'Onofrio, A. et al. Siderophores from neighboring organisms promote the growth of uncultured bacteria. Chem. Biol. 17, 254–264 (2010).
Human Microbiome Jumpstart Reference Strains Consortium. A catalog of reference genomes from the human microbiome. Science 328, 994–999 (2010). This paper presents methods and analysis for large-scale production of reference genome sequences from human-microbiome organisms.
Gans, J., Wolinsky, M. & Dunbar, J. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 309, 1387–1390 (2005).
Nelson, T. A. et al. PhyloChip microarray analysis reveals altered gastrointestinal microbial communities in a rat model of colonic hypersensitivity. Neurogastroenterol. Motil. 23, 169–177 (2011).
Bent, S. J. et al. Measuring species richness based on microbial community fingerprints: the emperor has no clothes. Appl. Environ. Microbiol. 73, 2399–2401 (2007).
Sogin, M. L. et al. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl Acad. Sci. USA 103, 12115–12120 (2006).
The NIH HMP Working Group et al. The NIH Human Microbiome Project. Genome Res. 19, 2317–2323 (2009).
Human Microbiome Project Consortium. A framework for human microbiome research. Nature 486, 215–221 (2012). This paper describes the data sets and resources of the HMP.
Lazarevic, V. et al. Metagenomic study of the oral microbiota by Illumina high-throughput sequencing. J. Microbiol. Methods 79, 266–271 (2009).
Claesson, M. J. et al. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 38, e200 (2010).
Gloor, G. B. et al. Microbiome profiling by Illumina sequencing of combinatorial sequence-tagged PCR products. PLoS ONE 5, e15406 (2010).
Hummelen, R. et al. Deep sequencing of the vaginal microbiota of women with HIV. PLoS ONE 5, e12078 (2010).
Zubrzycki, L. & Spaulding, E. H. Studies on the stability of the normal human fecal flora. J. Bacteriol. 83, 968–974 (1962).
Luckey, T. D. Introduction to intestinal microecology. Am. J. Clin. Nutr. 25, 1292–1294 (1972).
Arumugam, M. et al. Enterotypes of the human gut microbiome. Nature 473, 174–180 (2011).
Martin, J. et al. Optimizing read mapping to reference genomes to determine composition and species prevalence in microbial communities. PloS ONE 7, e36427 (2012).
Wu, G. D. et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108 (2011).
Ravel, J. et al. Vaginal microbiome of reproductive-age women. Proc. Natl Acad. Sci. USA 108, S4680–S4687 (2011).
Proctor, L. M. The Human Microbiome Project in 2011 and beyond. Cell Host Microbe 10, 287–291 (2011).
DOE Joint Genome Institute. A Genomic Encyclopedia of Bacteria and Archaea. http://www.jgi.doe.gov/programs/GEBA/ (US Department of Energy, 2012).
Parfrey, L. W., Walters, W. A. & Knight, R. Microbial eukaryotes in the human microbiome: ecology, evolution, and future directions. Front. Microbiol. 2, 153 (2011).
Wylie, K. M., Weinstock, G. M. & Storch, G. A. Emerging view of the human virome. Transl. Res. http://dx.doi.org/10.1016/j.trsl.2012.03.006 (24 April 2012).
Breitbart, M. et al. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 185, 6220–6223 (2003).
Palacios, G. et al. Panmicrobial oligonucleotide array for diagnosis of infectious diseases. Emerg. Infect. Dis. 13, 73–81 (2007).
Wang, D. et al. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol. 1, E2 (2003).
Casas, V. & Rohwer, F. Phage metagenomics. Methods Enzymol. 421, 259–268 (2007).
Allander, T. et al. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl Acad. Sci. USA 102, 12891–12896 (2005).
Finkbeiner, S. R. et al. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathogens. 4, e1000011 (2008).
Breitbart, M. & Rohwer, F. Method for discovering novel DNA viruses in blood using viral particle selection and shotgun sequencing. Biotechniques 39, 729–736 (2005).
Wylie, K. M., Mihindukulasuriya, K. A., Sodergren, E., Weinstock, G. M. & Storch, G. A. Sequence analysis of the human virome in febrile and afebrile children. PLoS ONE 7, e27735 (2012).
Pride, D. T. et al. Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J. 6, 915–926 (2011).
Minot, S., Grunberg, S., Wu, G. D., Lewis, J. D. & Bushman, F. D. Hypervariable loci in the human gut virome. Proc. Natl Acad. Sci. USA 109, 3962–3966 (2012).
Breitbart, M. et al. Viral diversity and dynamics in an infant gut. Res. Microbiol. 159, 367–373 (2008).
Minot, S. et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 21, 1616–1625 (2011).
Willner, D. & Furlan, M. Deciphering the role of phage in the cystic fibrosis airway. Virulence 1, 309–313 (2010).
Lepage, P. et al. Dysbiosis in inflammatory bowel disease: a role for bacteriophages? Gut 57, 424–425 (2008).
Kanehisa, M., Goto, S., Furumichi, M., Tanabe, M. & Hirakawa, M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 38, D355–D360 (2010).
Abubucker, S. et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput. Biol. 8, e1002358 (2012).
Cantarel, B. L. et al. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238 (2009).
Cantarel, B. L., Lombard, V. & Henrissat, B. Complex carbohydrate utilization by the healthy human microbiome. PLoS ONE 7, e28742 (2012).
Turnbaugh, P. J. & Gordon, J. I. The core gut microbiome, energy balance and obesity. J. Physiol. (Lond.) 587, 4153–4158 (2009).
Raes, J., Foerstner, K. U. & Bork, P. Get the most out of your metagenome: computational analysis of environmental sequence data. Curr. Opin. Microbiol. 10, 490–498 (2007).
Wooley, J. C., Godzik, A. & Friedberg, I. A primer on metagenomics. PLoS Comput. Biol. 6, e1000667 (2010).
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C. & Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200 (2011).
Haas, B. J. et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504 (2011).
Jumpstart Consortium Human Microbiome Project Data Generation Working Group. Evaluation of 16S rDNA-based community profiling for human microbiome research. PLoS ONE 7, e39315 (2012).
Schloss, P. D., Gevers, D. & Westcott, S. L. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS ONE 6, e27310 (2011).
Wright, E. S., Yilmaz, L. S. & Noguera, D. R. DECIPHER, a search-based approach to chimera identification for 16S rRNA sequences. Appl. Environ. Microbiol. 78, 717–725 (2012).
Lozupone, C., Lladser, M. E., Knights, D., Stombaugh, J. & Knight, R. UniFrac: an effective distance metric for microbial community comparison. ISME J. 5, 169–172 (2011).
Schloss, P. D. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009).
Caporaso, J. G. et al. Moving pictures of the human microbiome. Genome Biol. 12, R50 (2011).
Angiuoli, S. V., White, J. R., Matalka, M., White, O. & Fricke, W. F. Resources and costs for microbial sequence analysis evaluated using virtual machines and cloud computing. PLoS ONE 6, e26624 (2011).
Chitsaz, H. et al. Efficient de novo assembly of single-cell bacterial genomes from short-read data sets. Nature Biotechnol. 29, 915–921 (2011).
Dichosa, A. E. et al. Artificial polyploidy improves bacterial single cell genome recovery. PLoS ONE 7, e37387 (2012).
Benson, A. K. et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl Acad. Sci. USA 107, 18933–18938 (2010).
Elinav, E. et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 145, 745–757 (2011).
Hooper, L. V., Littman, D. R. & Macpherson, A. J. Interactions between the microbiota and the immune system. Science 336, 1268–1273 (2012).
Handelsman, J., Rondon, M. R., Brady, S. F., Clardy, J. & Goodman, R. M. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem. Biol. 5, R245–R249 (1998).
Riesenfeld, C. S., Schloss, P. D. & Handelsman, J. Metagenomics: genomic analysis of microbial communities. Annu. Rev. Genet. 38, 525–552 (2004).
Hooper, L. V. & Gordon, J. I. Commensal host–bacterial relationships in the gut. Science 292, 1115–1118 (2001).
Acknowledgements
The author gratefully acknowledges generous support from the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no competing financial interests.
Additional information
Reprints and permissions information is available at www.nature.com/reprints.
Rights and permissions
About this article
Cite this article
Weinstock, G. Genomic approaches to studying the human microbiota. Nature 489, 250–256 (2012). https://doi.org/10.1038/nature11553
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11553
This article is cited by
-
Pathophysiology of acute lung injury in patients with acute brain injury: the triple-hit hypothesis
Critical Care (2024)
-
Metagenomic sequencing reveals altered gut microbial compositions and gene functions in patients with non-segmental vitiligo
BMC Microbiology (2023)
-
In vitro gut microbiome response to carbohydrate supplementation is acutely affected by a sudden change in diet
BMC Microbiology (2023)
-
Characteristics of the oropharyngeal microbiota among infants with pneumonia and their effects on immune response and subsequent respiratory morbidity
European Journal of Pediatrics (2023)
-
Possible Role of Dysbiosis of the Gut Microbiome in SLE
Current Rheumatology Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.