Abstract
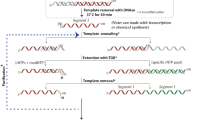
This protocol describes a method for direct labeling and detection of small RNAs present in total RNA by splinted ligation. The assay uses a small RNA-specific bridge oligonucleotide to form base pairs with the small RNA and a 5′-end-radiolabeled ligation oligonucleotide. The captured small RNA is directly labeled by ligation. Detection of the labeled small RNAs is performed by denaturing gel electrophoresis and autoradiography or phosphorimaging. This protocol has been successfully used to study expression of various classes of biological small RNAs from nanogram to microgram amounts of total RNA without an amplification step. It is significantly simpler to perform and more sensitive than either northern blotting or ribonuclease protection assays. Once the oligonucleotides have been synthesized and total RNA has been extracted, the procedure can be completed in 6 h.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Bartel, D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004).
Pillai, R.S. MicroRNA function: multiple mechanisms for a tiny RNA? RNA 11, 1753–1761 (2005).
Shingara, J. et al. An optimized isolation and labeling platform for accurate microRNA expression profiling. RNA 11, 1461–1470 (2005).
Hüttenhofer, A. & Vogel, J. Experimental approaches to identify non-coding RNAs. Nucleic Acids Res. 34, 635–646 (2006).
Chen, C. et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 33, e179 (2005).
Jiang, J., Lee, E.J., Gusev, Y. & Schmittgen, T.D. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res. 33, 5394–5403 (2005).
Jonstrup, S.P., Koch, J. & Kjems, J. A microRNA detection system based on padlock probes and rolling circle amplification. RNA 12, 1747–1752 (2006).
Moore, M.J. & Query, C.C. Joining of RNAs by splinted ligation. Methods Enzymol. 317, 109–123 (2000).
Maroney, P.A., Chamnongpol, S., Souret, F. & Nilsen, T.W. A rapid, quantitative assay for direct detection of microRNAs and other small RNAs using splinted ligation. RNA 13, 930–936 (2007).
Maroney, P.A., Yu, Y., Fisher, J. & Nilsen, T.W. Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat. Struct. Mol. Biol. 13, 1102–1107 (2006).
Sambrook, J. & Russell, D.W. Molecular Cloning: A Laboratory Manual. Chapter 7 Extraction, purification, and analysis of mRNA from eukaryotic cells (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 2001).
Acknowledgements
We thank Jesse Fisher for technical assistance, and Dr. John W. Chase and Christopher J. Kubu for helpful discussions.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
Case Western Reserve University is assigned intellectual property rights related to the described technology that have been and continue to be licensed in an exclusive manner to USB Corporation. Chamnongpol and Souret declare employment at USB Corporation.
Rights and permissions
About this article
Cite this article
Maroney, P., Chamnongpol, S., Souret, F. et al. Direct detection of small RNAs using splinted ligation. Nat Protoc 3, 279–287 (2008). https://doi.org/10.1038/nprot.2007.530
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2007.530
This article is cited by
-
A carbon dot and molecular beacon based fluorometric sensor for the cancer marker microRNA-21
Microchimica Acta (2019)
-
Heme enables proper positioning of Drosha and DGCR8 on primary microRNAs
Nature Communications (2017)
-
Fluorescence based turn-on strategy for determination of microRNA-155 using DNA-templated copper nanoclusters
Microchimica Acta (2017)
-
Changes in cellular microRNA expression induced by porcine circovirus type 2-encoded proteins
Veterinary Research (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.