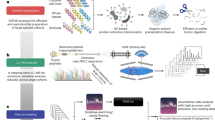
Abstract
In the analysis of biological systems, it is of interest to identify the components of the system and to monitor their changes in abundance under different conditions. The AQUA (for 'absolute quantification') method allows sensitive and specific targeted quantification of protein and post-translational modifications in complex protein mixtures using stable isotope–labeled peptides as internal standards. Each AQUA experiment is composed of two stages: method development and application to a biological scenario. In the method development stage, peptides from the protein of interest are chosen and then synthesized with stable isotopes such as 13C, 2H or 15N. The abundance of these internal standards and their endogenous counterparts can be measured by mass spectrometry with selected reaction monitoring or selected ion monitoring methods. Once an AQUA method is established, it can be rapidly applied to a wide range of biological samples, from tissue culture cells to human plasma and tissue. After AQUA peptide synthesis, the development, optimization and application of AQUA analyses to a specific biological problem can be achieved in ∼1 week. Here we demonstrate the usefulness of this method by monitoring both Polo-like kinase 1 (Plk1) protein abundance in multiple lung cancer cell lines and the extent of Plk1 activation loop phosphorylation (pThr-210) during release from S phase.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Ong, S.E. et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteomics 1, 376–86 (2002).
Gygi, S.P. et al. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 17, 994–9 (1999).
Ross, P.L. et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 (2004).
Gerber, S.A., Rush, J., Stemman, O., Kirschner, M.W. & Gygi, S.P. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. USA 100, 6940–6945 (2003).
Beynon, R.J., Doherty, M.K., Pratt, J.M. & Gaskell, S.J. Multiplexed absolute quantification in proteomics using artificial QCAT proteins of concatenated signature peptides. Nat. Methods 2, 587–589 (2005).
Pratt, J.M. et al. Multiplexed absolute quantification for proteomics using concatenated signature peptides encoded by QconCAT genes. Nat. Protoc. 1, 1029–1043 (2006).
Langenfeld, E., Zanger, U.M., Jung, K., Meyer, H.E. & Marcus, K. Mass spectrometry-based absolute quantification of microsomal cytochrome P450 2D6 in human liver. Proteomics 9, 2313–2323 (2009).
Mayya, V., Rezual, K., Wu, L., Fong, M.B. & Han, D.K. Absolute quantification of multisite phosphorylation by selective reaction monitoring mass spectrometry: determination of inhibitory phosphorylation status of cyclin-dependent kinases. Mol. Cell. Proteomics 5, 1146–1157 (2006).
Kirkpatrick, D.S. et al. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat. Cell Biol. 8, 700–710 (2006).
Xu, P. et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 137, 133–145 (2009).
Deutsch, E.W., Lam, H. & Aebersold, R. PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 9, 429–434 (2008).
Mathivanan, S. et al. Human Proteinpedia enables sharing of human protein data. Nat. Biotechnol. 26, 164–167 (2008).
Jones, P. et al. PRIDE: a public repository of protein and peptide identifications for the proteomics community. Nucleic Acids Res. 34, D659–D663 (2006).
Mallick, P. et al. Computational prediction of proteotypic peptides for quantitative proteomics. Nat. Biotechnol. 25, 125–131 (2007).
Picotti, P. et al. High-throughput generation of selected reaction-monitoring assays for proteins and proteomes. Nat. Methods 7, 43–46 (2010).
MacLean, B. et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 (2010).
Papaioannou, M.D. et al. Loss of Dicer in Sertoli cells has a major impact on the testicular proteome of mice. Mol. Cell Proteomics Published online; doi:10.1074/mcp.M900587-MCP200 (13 May 2010).
Stemmann, O., Zou, H., Gerber, S.A., Gygi, S.P. & Kirschner, M.W. Dual inhibition of sister chromatid separation at metaphase. Cell 107, 715–726 (2001).
Wolf, G. et al. Prognostic significance of polo-like kinase (PLK) expression in non-small cell lung cancer. Oncogene 14, 543–549 (1997).
Seki, A., Coppinger, J.A., Jang, C.Y., Yates, J.R. & Fang, G. Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science 320, 1655–1658 (2008).
Macurek, L. et al. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature 455, 119–123 (2008).
Olsen, J.V. et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3, ra3 (2010).
Beausoleil, S.A. et al. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. USA 101, 12130–12135 (2004).
Acknowledgements
We thank S. Cullati and D. Schweppe for helpful discussion and comments on the manuscript. This work was supported by the National Institutes of Health Grant P20-RR018787 from the IDeA Program of the National Center for Research Resources and the American Cancer Society Grant IRG-82-003-24 (to S.A.G.).
Author information
Authors and Affiliations
Contributions
A.N.K. designed and conducted the experiments for Plk1 AQUA analysis, interpreted the data and wrote the paper; J.R. synthesized the Plk1 AQUA peptides used in this study; and S.A.G. conceived of the experiments for Plk1 AQUA analysis, interpreted the data and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
Scott Gerber is a patent holder for AQUA technology, and receives royalty income from this patent. John Rush is an employee of Cell Signaling Technology, which has licensed the AQUA technology and sells AQUA peptides.
Rights and permissions
About this article
Cite this article
Kettenbach, A., Rush, J. & Gerber, S. Absolute quantification of protein and post-translational modification abundance with stable isotope–labeled synthetic peptides. Nat Protoc 6, 175–186 (2011). https://doi.org/10.1038/nprot.2010.196
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2010.196
This article is cited by
-
Sequential phosphoproteomics and N-glycoproteomics of plasma-derived extracellular vesicles
Nature Protocols (2020)
-
Accurate and easy method for systemin quantification and examining metabolic changes under different endogenous levels
Plant Methods (2018)
-
Protein-species quantitative venomics: looking through a crystal ball
Journal of Venomous Animals and Toxins including Tropical Diseases (2017)
-
A quantitative analytical method for PIVKA-II using multiple reaction monitoring-mass spectrometry for early diagnosis of hepatocellular carcinoma
Analytical and Bioanalytical Chemistry (2017)
-
Characterization of SNPs in the dopamine-β-hydroxylase gene providing new insights into its structure-function relationship
neurogenetics (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.