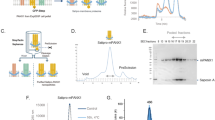
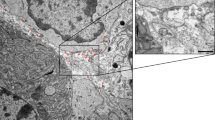
Abstract
Dynamic interplay between intracellular organelles requires a particular functional apposition of membrane structures. The organelles involved come into close contact, but do not fuse, thereby giving rise to notable microdomains; these microdomains allow rapid communication between the organelles. Plasma membrane–associated membranes (PAMs), which are microdomains of the plasma membrane (PM) interacting with the endoplasmic reticulum (ER) and mitochondria, are dynamic structures that mediate transport of proteins, lipids, ions and metabolites. These structures have gained much interest lately owing to their roles in many crucial cellular processes. Here we provide an optimized protocol for the isolation of PAM, PM and ER fractions from rat liver that is based on a series of differential centrifugations, followed by the fractionation of crude PM on a discontinuous sucrose gradient. The procedure requires ∼8–10 h, and it can be easily modified and adapted to other tissues and cell types.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Pichler, H. et al. A subfraction of the yeast endoplasmic reticulum associates with the plasma membrane and has a high capacity to synthesize lipids. Eur. J. Biochem. 268, 2351–2361 (2001).
Achleitner, G. et al. Association between the endoplasmic reticulum and mitochondria of yeast facilitates interorganelle transport of phospholipids through membrane contact. Eur. J. Biochem. 264, 545–553 (1999).
Wu, M.M., Buchanan, J., Luik, R.M. & Lewis, R.S. Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J. Cell Biol. 174, 803–813 (2006).
Lebiedzinska, M., Duszynski, J., Rizzuto, R., Pinton, P. & Wieckowski, M.R. Age-related changes in levels of p66Shc and serine 36-phosphorylated p66Shc in organs and mouse tissues. Arch. Biochem. Biophys. 486, 73–80 (2009).
Vance, J.E. & Tasseva, G. Formation and function of phosphatidylserine and phosphatidylethanolamine in mammalian cells. Biochim. Biophys. Acta. 1831, 543–554 (2013).
Perry, R.J. & Ridgway, N.D. Oxysterol-binding protein and vesicle-associated membrane protein-associated protein are required for sterol-dependent activation of the ceramide transport protein. Mol. Biol. Cell 17, 2604–2616 (2006).
Yan, D. & Olkkonen, V.M. Characteristics of oxysterol binding proteins. Int. Rev. Cytol. 265, 253–285 (2008).
Kozieł, K. et al. Plasma membrane associated membranes (PAM) from Jurkat cells contain STIM1 protein: is PAM involved in the capacitative calcium entry? Int. J. Biochem. Cell Biol. 41, 2440–2449 (2009).
Camici, O. & Corazzi, L. Phosphatidylserine translocation into brain mitochondria: involvement of a fusogenic protein associated with mitochondrial membranes. Mol. Cell Biochem. 175, 71–80 (1997).
Lapierre, L.A., Tuma, P.L., Navarre, J., Goldenring, J.R. & Anderson, J.M. VAP-33 localizes to both an intracellular vesicle population and with occludin at the tight junction. J. Cell Sci. 112, 3723–3732 (1999).
Lebiedzinska, M., Szabadkai, G., Jones, A.W., Duszynski, J. & Wieckowski, M.R. Interactions between the endoplasmic reticulum, mitochondria, plasma membrane and other subcellular organelles. Int. J. Biochem. Cell Biol. 41, 1805–1816 (2009).
Kakizawa, S., Moriguchi, S., Ikeda, A., Iino, M. & Takeshima, H. Functional crosstalk between cell-surface and intracellular channels mediated by junctophilins essential for neuronal functions. Cerebellum 7, 385–391 (2008).
Anderie, I., Schulz, I. & Schmid, A. Direct interaction between ER membrane-bound PTP1B and its plasma membrane-anchored targets. Cell Signal 19, 582–592 (2007).
Goldstein, B.J., Bittner-Kowalczyk, A., White, M.F. & Harbeck, M. Tyrosine dephosphorylation and deactivation of insulin receptor substrate-1 by protein-tyrosine phosphatase 1B. Possible facilitation by the formation of a ternary complex with the Grb2 adaptor protein. J. Biol. Chem. 275, 4283–4289 (2000).
Hernández, M.V., Sala, M.G., Balsamo, J., Lilien, J. & Arregui, C.O. ER-bound PTP1B is targeted to newly forming cell-matrix adhesions. J. Cell Sci. 119, 1233–1243 (2006).
Helle, S.C. et al. Organization and function of membrane contact sites. Biochim. Biophys. Acta 1833, 2526–2541 (2013).
Leitman, J., Ron, E., Ogen-Shtern, N. & Lederkremer, G.Z. Compartmentalization of endoplasmic reticulum quality control and ER-associated degradation factors. DNA Cell Biol. 32, 2–7 (2013).
Lam, A.K. & Galione, A. The endoplasmic reticulum and junctional membrane communication during calcium signaling. Biochim. Biophys. Acta 1833, 2542–2559 (2013).
Quintana, A. et al. Sustained activity of calcium release-activated calcium channels requires translocation of mitochondria to the plasma membrane. J. Biol. Chem. 281, 40302–40309 (2006).
Poburko, D. et al. Mitochondria buffer NCX-mediated Ca2+-entry and limit its diffusion into vascular smooth muscle cells. Cell Calcium 40, 359–371 (2006).
Giorgi, C., De Stefani, D., Bononi, A., Rizzuto, R. & Pinton, P. Structural and functional link between the mitochondrial network and the endoplasmic reticulum. Int. J. Biochem. Cell Biol. 41, 1817–1827 (2009).
Wieckowski, M.R., Giorgi, C., Lebiedzinska, M., Duszynski, J. & Pinton, P. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat. Protoc. 4, 1582–1590 (2009).
England, M.D. et al. Influence of antioxidants (mannitol and allopurinol) on oxygen free radical generation during and after cardiopulmonary bypass. Circulation 74, 134–137 (1986).
Nur, T., Sela, I., Webster, N.J. & Madar, Z. Starvation and refeeding regulate glycogen synthase gene expression in rat liver at the posttranscriptional level. J. Nutr. 125, 2457–2462 (1995).
Gold, L.I. et al. Calreticulin: non-endoplasmic reticulum functions in physiology and disease. FASEB J. 24, 665–683 (2010).
Lillemeier, B.F., Pfeiffer, J.R., Surviladze, Z., Wilson, B.S. & Davis, M.M. Plasma membrane-associated proteins are clustered into islands attached to the cytoskeleton. Proc. Natl. Acad. Sci. USA 103, 18992–18997 (2006).
Hinton, R.H. Purification of plasma membrane fragments. In Subcellular components. Preparation and fractionation 2nd edn, (ed. Birnie, G.D.) 119–156 (Butterworths, 1972).
Acknowledgements
This research was supported by grants from the Polish National Science Centre (UMO-2011/01/M/NZ3/02128), Iuventus Plus (UMO-0531/IP1/2011/71) and Polish Ministry of Science and Higher Education grant W100/HFSC/2011 to J.M.S., M.L., A.W., J.D. and M.R.W.; by the Italian Association for Cancer Research to C.G. and P.P.; and by Telethon (GGP11139B), the Italian Ministry of Education, University and Research, and the Italian Ministry of Health to P.P. J.M.S. was also supported by a Ph.D. fellowship from The Foundation for Polish Science (FNP), and by the EU, the European Regional Development Fund and the 'Innovative economy' Operational Programme. M.L. was supported by the 'Mobility Plus' fellowship from the Polish Ministry of Science and Higher Education.
Author information
Authors and Affiliations
Contributions
J.M.S., M.L., A.W. and C.G. carried out all experimental steps and performed characterization of the isolated subcellular fractions. J.M.S., J.D., P.P. and M.R.W. developed the concept, designed experiments and wrote the paper. All authors discussed the results and commented on the manuscript at all stages.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Suski, J., Lebiedzinska, M., Wojtala, A. et al. Isolation of plasma membrane–associated membranes from rat liver. Nat Protoc 9, 312–322 (2014). https://doi.org/10.1038/nprot.2014.016
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2014.016
This article is cited by
-
A chemical proteomics approach for global mapping of functional lysines on cell surface of living cell
Nature Communications (2024)
-
Cell membrane-based nanomaterials for theranostics of central nervous system diseases
Journal of Nanobiotechnology (2023)
-
Spectrin-beta 2 facilitates the selective accumulation of GABAA receptors at somatodendritic synapses
Communications Biology (2023)
-
Disruption of ER ion homeostasis maintained by an ER anion channel CLCC1 contributes to ALS-like pathologies
Cell Research (2023)
-
Neutrophil membrane engineered HucMSC sEVs alleviate cisplatin-induced AKI by enhancing cellular uptake and targeting
Journal of Nanobiotechnology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.