Key Points
-
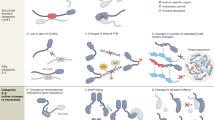
Alternative splicing allows individual genes to generate multiple mRNAs. Many of these mRNAs encode functionally distinct protein isoforms, thereby bridging the gap between genome and proteome.
-
Alternative splicing mechanisms have traditionally been investigated using individual model systems, but these approaches are now being complemented by global analyses. The regulation of splicing commonly involves the modulation of early steps in spliceosome assembly. The cis elements comprise splicing enhancers and silencers that can be located in either the exons or the introns and that bind activator and repressor proteins.
-
Sometimes, the presence or absence of a single regulator is sufficient to determine alternative splicing pathways. More commonly, combinations of more widespread factors are involved in the choice of splicing pathway. This gives rise to the concept of 'cellular codes', which are constituted by particular combinations of regulatory factors.
-
Members of the SR protein family can activate splicing by binding to exon splicing enhancers (ESEs). Models for the action of enhancers include the recruitment of the U2 auxiliary factor (U2AF) to weak 3′ splice sites, the interaction with co-activators and direct contact with the RNA at the branch point.
-
Repressors, which are frequently members of the heterogeneous nuclear ribonucleoprotein (hnRNP) family, function by making splice sites inaccessible or by promoting the formation of 'dead-end' splicing related complexes. Alternative splicing decisions frequently involve the dynamic antagonism between regulatory activators and repressors.
-
Several global strategies for identifying splicing silencers and enhancers have been successfully deployed. These include RNA-binding SELEX studies, functional SELEX in vitro, cell-based selection assays and computational surveys for the differential enrichment of sequence motifs in expected locations for silencers and enhancers.
-
Methods are being developed to detect complete sets of cellular RNAs with which individual splicing regulators interact. These include assays that detect RNAs to which the factor binds, or alternative splicing events that are affected by depletion of the regulator. A key technical development is the recent introduction of alternative splicing microarrays, which allow parallel analysis of large numbers of alternative splicing events and should facilitate the deciphering of cellular codes.
Abstract
In violation of the 'one gene, one polypeptide' rule, alternative splicing allows individual genes to produce multiple protein isoforms — thereby playing a central part in generating complex proteomes. Alternative splicing also has a largely hidden function in quantitative gene control, by targeting RNAs for nonsense-mediated decay. Traditional gene-by-gene investigations of alternative splicing mechanisms are now being complemented by global approaches. These promise to reveal details of the nature and operation of cellular codes that are constituted by combinations of regulatory elements in pre-mRNA substrates and by cellular complements of splicing regulators, which together determine regulated splicing pathways.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Burge, C., Tuschl, T. & Sharp, P. in The RNA World 2nd edn (eds Gestetland, R., Cech, T. & Atkins, J.) 525–560 (Cold Spring Harbor Laboratory Press, 1999).
Jurica, M. S. & Moore, M. J. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 12, 5–14 (2003).
Cartegni, L., Chew, S. L. & Krainer, A. R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nature Rev. Genet. 3, 285–298 (2002).
Caceres, J. F. & Kornblihtt, A. R. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet. 18, 186–193 (2002).
Faustino, N. A. & Cooper, T. A. Pre-mRNA splicing and human disease. Genes Dev. 17, 419–437 (2003).
Pagani, F. & Baralle, F. E. Genomic variants in exons and introns: identifying the splicing spoilers. Nature Rev. Genet. 5, 389–396 (2004).
Modrek, B. & Lee, C. A genomic view of alternative splicing. Nature Genet. 30, 13–19 (2002).
Johnson, J. M. et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302, 2141–2144 (2003). Used a splice-sensitive microarray with probes for every known exon–exon junction in >10,000 human genes to reveal several global features of alternative splicing, as well as the discovery and validation of previously uncharacterized splicing events.
Kampa, D. et al. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res. 14, 331–342 (2004).
Thanaraj, T. A., Clark, F. & Muilu, J. Conservation of human alternative splice events in mouse. Nucleic Acids Res. 31, 2544–2552 (2003).
Pan, Q. et al. Alternative splicing of conserved exons is frequently species-specific in human and mouse. Trends Genet. 277, 73–77 (2005).
Nurtdinov, R. N., Artamonova, I. I., Mironov, A. A. & Gelfand, M. S. Low conservation of alternative splicing patterns in the human and mouse genomes. Hum. Mol. Genet. 12, 1313–1320 (2003).
Modrek, B. & Lee, C. J. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nature Genet. 34, 177–180 (2003).
Philipps, D. L., Park, J. W. & Graveley, B. R. A computational and experimental approach toward a priori identification of alternatively spliced exons. RNA 10, 1838–1844 (2004).
Sorek, R. & Ast, G. Intronic sequences flanking alternatively spliced exons are conserved between human and mouse. Genome Res. 13, 1631–1637 (2003).
Sorek, R. et al. A non-EST-based method for exon-skipping prediction. Genome Res. 14, 1617–1623 (2004).
Clark, F. & Thanaraj, T. A. Categorization and characterization of transcript-confirmed constitutively and alternatively spliced introns and exons from human. Hum. Mol. Genet. 11, 451–464 (2002).
Lewis, B. P., Green, R. E. & Brenner, S. E. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl Acad. Sci. USA 100, 189–192 (2003). A bioinformatics survey indicating that a large proportion of human alternative splicing leads to products that are degraded by nonsense-mediated decay.
Schmucker, D. & Flanagan, J. G. Generation of recognition diversity in the nervous system. Neuron 44, 219–222 (2004).
Wojtowicz, W. M., Flanagan, J. J., Millard, S. S., Zipursky, S. L. & Clemens, J. C. Alternative splicing of Drosophila Dscam generates axon guidance receptors that exhibit isoform-specific homophilic binding. Cell 118, 619–633 (2004).
Lopez, A. J. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu. Rev. Genet. 32, 279–305 (1998).
Jensen, K. B. et al. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron 25, 359–371 (2000). The mouse knockout revealed Nova1 to be a mammalian tissue-specific splicing regulator.
Black, D. L. Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell 103, 367–370 (2000).
Smith, C. W. & Valcarcel, J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 25, 381–388 (2000).
Shin, C. & Manley, J. L. Cell signalling and the control of pre-mRNA splicing. Nature Rev. Mol. Cell Biol. 5, 727–738 (2004).
Stamm, S. Signals and their transduction pathways regulating alternative splicing: a new dimension of the human genome. Hum. Mol. Genet. 11, 2409–2416 (2002).
Ast, G. How did alternative splicing evolve? Nature Rev. Genet. 5, 773–782 (2004).
Wang, Z. et al. Systematic identification and analysis of exonic splicing silencers. Cell 119, 831–845 (2004). A transfection-based screen for ESSs, which can potentially be used to identify different classes of regulatory element that are active in different cell types.
Zhang, X. H. & Chasin, L. A. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 18, 1241–1250 (2004). A computational screen for ESSs and ESEs; the datasets produced by reference 28 and 29 allow pseudo-exons to be discriminated from authentic exons.
Graveley, B. R. Sorting out the complexity of SR protein functions. RNA 6, 1197–1211 (2000).
Ring, H. Z. & Lis, J. T. The SR protein B52/SRp55 is essential for Drosophila development. Mol. Cell. Biol. 14, 7499–7506 (1994).
Xu, X. et al. ASF/SF2-regulated CaMKIId alternative splicing temporally reprograms excitation–contraction coupling in cardiac muscle. Cell 120, 59–72 (2005). Conditional Cre-mediated knockout of SF2/ASF in cardiac muscle produces a surprisingly specific phenotype with disruption of a few specific alternative splicing events.
Wang, J., Takagaki, Y. & Manley, J. L. Targeted disruption of an essential vertebrate gene: ASF/SF2 is required for cell viability. Genes Dev. 10, 2588–2599 (1996).
Jumaa, H., Wei, G. & Nielsen, P. J. Blastocyst formation is blocked in mouse embryos lacking the splicing factor SRp20. Curr. Biol. 9, 899–902 (1999).
Ding, J. H. et al. Dilated cardiomyopathy caused by tissue-specific ablation of SC35 in the heart. EMBO J. 23, 885–896 (2004).
Shen, H. & Green, M. R. A pathway of sequential arginine-serine-rich domain-splicing signal interactions during mammalian spliceosome assembly. Mol. Cell 16, 363–373 (2004).
Shen, H., Kan, J. L. & Green, M. R. Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol. Cell 13, 367–376 (2004). References 36 and 37 show specific interactions of RS domains with the branch point and 5′ splice site sequences within splicing complexes, which indicates that the key functional role of RS domains might be to bind RNA.
Krecic, A. M. & Swanson, M. S. hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol. 11, 363–371 (1999).
Dreyfuss, G., Kim, V. K. & Kataoka, N. Messenger RNA-binding proteins and the messages they carry. Nature Rev. Mol. Cell Biol. 3, 195–205 (2002).
Bennett, M., Pinol-Roma, S., Staknis, D., Dreyfuss, G. & Reed, R. Differential binding of heterogeneous nuclear ribonucleoproteins to mRNA precursors prior to spliceosome assembly in vitro. Mol. Cell. Biol. 12, 3165–3175 (1992).
Buratti, E. & Baralle, F. E. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol. Cell. Biol. 24, 10505–10514 (2004).
Smith, C. W. J. & Nadal Ginard, B. Mutually exclusive splicing of α-tropomyosin exons enforced by an unusual lariat branch point location: implications for constitutive splicing. Cell 56, 749–758 (1989).
Kornblihtt, A. R., de la Mata, M., Fededa, J. P., Munoz, M. J. & Nogues, G. Multiple links between transcription and splicing. RNA 10, 1489–1498 (2004).
Maniatis, T. & Reed, R. An extensive network of coupling among gene expression machines. Nature 416, 499–506 (2002).
Lynch, K. W. & Maniatis, T. Synergistic interactions between two distinct elements of a regulated splicing enhancer. Genes Dev. 9, 284–293 (1995).
Lynch, K. W. & Maniatis, T. Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer. Genes Dev. 10, 2089–2101 (1996).
Tian, M. & Maniatis, T. A splicing enhancer exhibits both constitutive and regulated activities. Genes Dev. 8, 1703–1712 (1994).
Zuo, P. & Maniatis, T. The splicing factor U2AF35 mediates critical protein–protein interactions in constitutive and enhancer-dependent splicing. Genes Dev. 10, 1356–1368 (1996).
Rudner, D. Z., Breger, K. S. & Rio, D. C. Molecular genetic analysis of the heterodimeric splicing factor U2AF: the RS domain on either the large or small Drosophila subunit is dispensable in vivo. Genes Dev. 12, 1010–1021 (1998).
Kan, J. L. & Green, M. R. Pre-mRNA splicing of IgM exons M1 and M2 is directed by a juxtaposed splicing enhancer and inhibitor. Genes Dev. 13, 462–471 (1999).
Li, Y. & Blencowe, B. J. Distinct factor requirements for exonic splicing enhancer function and binding of U2AF to the polypyrimidine tract. J. Biol. Chem. 274, 35074–35079 (1999).
Eldridge, A. G., Li, Y., Sharp, P. A. & Blencowe, B. J. The SRm160/300 splicing co-activator is required for exon-enhancer function. Proc. Natl Acad. Sci. USA 96, 6125–6130 (1999).
Blencowe, B. J. et al. The SRm160/300 splicing co-activator subunits. RNA 6, 111–120 (2000).
Heinrichs, V., Ryner, L. C. & Baker, B. S. Regulation of sex-specific selection of fruitless 5′ splice sites by transformer and transformer-2. Mol. Cell. Biol. 18, 450–458 (1998).
Lam, B. J. et al. Enhancer-dependent 5′-splice site control of fruitless pre-mRNA splicing. J. Biol. Chem. 278, 22740–22747 (2003).
Wu, J. Y. & Maniatis, T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 75, 1061–1070 (1993). The original demonstration of specific interaction between RS domains, on which many subsequent models of SR protein action have been based.
Graveley, B. R. A protein interaction domain contacts RNA in the prespliceosome. Mol. Cell 13, 302–304 (2004).
Hui, J., Stangl, K., Lane, W. S. & Bindereif, A. HnRNP L stimulates splicing of the eNOS gene by binding to variable-length CA repeats. Nature Struct. Biol. 10, 33–37 (2003).
Del Gatto-Konczak, F. et al. The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol. Cell. Biol. 20, 6287–6299 (2000).
Forch, P. et al. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol. Cell. 6, 1089–1098 (2000).
Forch, P., Puig, O., Martinez, C., Seraphin, B. & Valcarcel, J. The splicing regulator TIA-1 interacts with U1-C to promote U1 snRNP recruitment to 5′ splice sites. EMBO J. 21, 6882–6892 (2002).
Le Guiner, C. et al. TIA-1 and TIAR activate splicing of alternative exons with weak 5′ splice sites followed by a U-rich stretch on their own pre-mRNAs. J. Biol. Chem. 276, 40638–40646 (2001).
Zhu, H., Hasman, R. A., Young, K. M., Kedersha, N. L. & Lou, H. U1 snRNP-dependent function of TIAR in the regulation of alternative RNA processing of the human calcitonin/CGRP pre-mRNA. Mol. Cell. Biol. 23, 5959–5971 (2003).
Wagner, E. J. & Garcia-Blanco, M. A. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 21, 3281–3288 (2001).
Valcarcel, J., Singh, R., Zamore, P. D. & Green, M. R. The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature 362, 171–175 (1993).
Lin, C. H. & Patton, J. G. Regulation of alternative 3′ splice site selection by constitutive splicing factors. RNA 1, 234–245 (1995).
Singh, R., Valcarcel, J. & Green, M. R. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 268, 1173–1176 (1995).
Zhang, L., Liu, W. & Grabowski, P. J. Coordinate repression of a trio of neuron-specific splicing events by the splicing regulator PTB. RNA 5, 117–130 (1999).
Southby, J., Gooding, C. & Smith, C. W. Polypyrimidine tract binding protein functions as a repressor to regulate alternative splicing of α-actinin mutally exclusive exons. Mol. Cell. Biol. 19, 2699–2711 (1999).
Chou, M. Y., Underwood, J. G., Nikolic, J., Luu, M. H. T. & Black, D. L. Multisite RNA binding and release of polypyrimidine tract binding protein during the regulation of c-src neural-specific splicing. Mol. Cell 5, 949–957 (2000). Using site-specifically labelled RNAs, the authors show cooperative binding of PTB to sites flanking the src N1 exon as the cause of exon skipping.
Wagner, E. J. & Garcia-Blanco, M. A. RNAi-mediated PTB depletion leads to enhanced exon definition. Mol. Cell 10, 943–949 (2002). Demonstrates, by RNAi and artificial recruitment, that PTB functions as a splicing repressor in vivo.
Wang, J. & Bell, L. R. The Sex-lethal amino terminus mediates cooperative interactions in RNA binding and is essential for splicing regulation. Genes Dev. 8, 2072–2085 (1994).
Lallena, M. J., Chalmers, K. J., Llamazares, S., Lamond, A. I. & Valcarcel, J. Splicing regulation at the second catalytic step by Sex-lethal involves 3′ splice site recognition by SPF45. Cell 109, 285–296 (2002). Shows that SXL represses splicing of its own exon 3, apparently by inhibiting splicing after the first catalytic step.
Mayeda, A. & Krainer, A. R. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell 68, 365–375 (1992). The first demonstration of antagonistic effects of hnRNPA1 and SF2/ASF upon alternative splicing.
Eperon, I. C. et al. Selection of alternative 5′ splice sites: role of U1 snRNP and models for the antagonistic effects of SF2/ASF and hnRNP A1. Mol. Cell. Biol. 20, 8303–8318 (2000).
Caceres, J. F., Stamm, S., Helfman, D. M. & Krainer, A. R. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science 265, 1706–1709 (1994).
van der Houven van Oordt, W. et al. The MKK(3/6)-p38-signaling cascade alters the subcellular distribution of hnRNP A1 and modulates alternative splicing regulation. J. Cell Biol. 149, 307–316 (2000). An important demonstration of a signalling pathway that alters alternative splicing patterns by relocalizing hnRNP A1 to the cytoplasm, thereby altering the nuclear ratio of hnRNP A1 and SF2/ASF.
Blanchette, M. & Chabot, B. Modulation of exon skipping by high-affinity hnRNP A1-binding sites and by intron elements that repress splice site utilization. EMBO J. 18, 1939–1952 (1999).
Zhu, J., Mayeda, A. & Krainer, A. R. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol. Cell 8, 1351–1361 (2001). Characterizes an ESE that functions by blocking the propagation of hnRNP A1 binding from a downstream ESS; in contrast to other ESEs, this one relies only on the RRMs and not the RS domain of bound SF2/ASF.
Del Gatto Konczak, F., Olive, M., Gesnel, M. C. & Breathnach, R. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol. Cell. Biol. 19, 251–260 (1999). Showed that an ESS functions by binding hnRNP A1, and that recruitment of just the glycine-rich domain of hnRNP A1 is sufficient for ESS function.
Tange, T. O., Damgaard, C. K., Guth, S., Valcarcel, J. & Kjems, J. The hnRNP A1 protein regulates HIV-1 tat splicing via a novel intron silencer element. EMBO J. 20, 5748–5758 (2001).
Villemaire, J., Dion, I., Elela, S. A. & Chabot, B. Reprogramming alternative pre-messenger RNA splicing through the use of protein-binding antisense oligonucleotides. J. Biol. Chem. 278, 50031–50039 (2003).
Baraniak, A. P., Lasda, E. L., Wagner, E. J. & Garcia-Blanco, M. A. A stem structure in fibroblast growth factor receptor 2 transcripts mediates cell-type-specific splicing by approximating intronic control elements. Mol. Cell. Biol. 23, 9327–9337 (2003).
Kanopka, A. et al. Regulation of adenovirus alternative RNA splicing by dephosphorylation of SR proteins. Nature 393, 185–187 (1998).
Dauksaite, V. & Akusjarvi, G. Human splicing factor ASF/SF2 encodes for a repressor domain required for its inhibitory activity on pre-mRNA splicing. J. Biol. Chem. 277, 12579–12586 (2002). Using artificial recruitment, the authors show that the second RRM domain of SF2/ASF functions as a splicing repressor when recruited upstream of a regulated 3′ splice site.
Estmer Nilsson, C. et al. The adenovirus E4-ORF4 splicing enhancer protein interacts with a subset of phosphorylated SR proteins. EMBO J. 20, 864–871 (2001).
Shen, H., Kan, J. L., Ghigna, C., Biamonti, G. & Green, M. R. A single polypyrimidine tract binding protein (PTB) binding site mediates splicing inhibition at mouse IgM exons M1 and M2. RNA 10, 787–794 (2004).
Labourier, E., Adams, M. D. & Rio, D. C. Modulation of P-element pre-mRNA splicing by a direct interaction between PSI and U1 snRNP 70K protein. Mol. Cell 8, 363–373 (2001). Shows a direct interaction between P-element somatic inhibitor and U1 snRNP 70K protein, which leads to splicing repression by diverting U1 binding to pseudo-5′ splice sites in the upstream exon.
Cote, J., Dupuis, S. & Wu, J. Y. Polypyrimidine track-binding protein binding downstream of caspase-2 alternative exon 9 represses its inclusion. J. Biol. Chem. 276, 8535–8543 (2001).
Jumaa, H. & Nielsen, P. J. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J. 16, 5077–5085 (1997).
Labourier, E. et al. Antagonism between RSF1 and SR proteins for both splice-site recognition in vitro and Drosophila development. Genes Dev. 13, 740–753 (1999).
Labourier, E. et al. Recognition of exonic splicing enhancer sequences by the Drosophila splicing repressor RSF1. Nucleic Acids Res. 27, 2377–2386 (1999).
Barnard, D. C. & Patton, J. G. Identification and characterization of a novel serine-arginine-rich splicing regulatory protein. Mol. Cell. Biol. 20, 3049–3057 (2000).
Li, J. et al. Regulation of alternative splicing by SRrp86 and its interacting proteins. Mol. Cell. Biol. 23, 7437–7447 (2003).
Ladd, A. N., Charlet-B., N. & Cooper, T. A. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol. Cell. Biol. 21, 1285–1296 (2001).
Charlet-B., N., Logan, P., Singh, G. & Cooper, T. A. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol. Cell 9, 649–658 (2002).
Zhang, W., Liu, H., Han, K. & Grabowski, P. J. Region-specific alternative splicing in the nervous system: implications for regulation by the RNA-binding protein NAPOR. RNA 8, 671–685 (2002).
Suzuki, H., Jin, Y., Otani, H., Yasuda, K. & Inoue, K. Regulation of alternative splicing of α-actinin transcript by Bruno-like proteins. Genes Cells 7, 133–141 (2002).
Gromak, N., Matlin, A. J., Cooper, T. A. & Smith, C. W. Antagonistic regulation of α-actinin alternative splicing by CELF proteins and polypyrimidine tract binding protein. RNA 9, 443–456 (2003).
Black, D. L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72, 291–336 (2003).
Markovtsov, V. et al. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell. Biol. 20, 7463–7479 (2000).
Rooke, N., Markovtsov, V., Cagavi, E. & Black, D. L. Roles for SR proteins and hnRNP A1 in the regulation of c-src exon N1. Mol. Cell. Biol. 23, 1874–1884 (2003).
Ule, J. et al. CLIP identifies Nova-regulated RNA networks in the brain. Science 302, 1212–1215 (2003). Reports a novel technique that allowed the identification of cellular targets of Nova proteins in mouse brain, many of which were subsequently validated by analysing their splicing patterns in Nova-knockout mice.
Polydorides, A. D., Okano, H. J., Yang, Y. Y., Stefani, G. & Darnell, R. B. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proc. Natl Acad. Sci. USA 97, 6350–6355 (2000).
Ladd, A. N. & Cooper, T. A. Finding signals that regulate alternative splicing in the post–genomic era. Genome Biol. 3, reviews 0008 (2002).
Zheng, Z. M. Regulation of alternative RNA splicing by exon definition and exon sequences in viral and mammalian gene expression. J. Biomed. Sci. 11, 278–294 (2004).
Tacke, R. & Manley, J. L. Determinants of SR protein specificity. Curr. Opin. Cell Biol. 11, 358–362 (1999).
Burd, C. G. & Dreyfuss, G. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 13, 1197–1204 (1994).
Perez, I., Lin, C. H., McAfee, J. G. & Patton, J. G. Mutation of PTB binding sites causes misregulation of alternative 3′ splice site selection in vivo. RNA 3, 764–778 (1997).
Jin, Y. et al. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 22, 905–912 (2003).
Brudno, M. et al. Computational analysis of candidate intron regulatory elements for tissue-specific alternative pre-mRNA splicing. Nucleic Acids Res. 29, 2338–2348 (2001).
Liu, H. X., Chew, S. L., Cartegni, L., Zhang, M. Q. & Krainer, A. R. Exonic splicing enhancer motif recognized by human SC35 under splicing conditions. Mol. Cell. Biol. 20, 1063–1071 (2000).
Liu, H. X., Zhang, M. & Krainer, A. R. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 12, 1998–2012 (1998). References 112 and 113 describe a functional SELEX approach in which ESEs were selected in cytoplasmic S100 extract in the presence of individual SR proteins, allowing the identification of ESEs specific for each protein. This allowed the development of the ESEfinder web resource.
Schaal, T. D. & Maniatis, T. Selection and characterization of pre-mRNA splicing enhancers: identification of novel SR protein-specific enhancer sequences. Mol. Cell. Biol. 19, 1705–1719 (1999).
Tian, H. & Kole, R. Selection of novel exon recognition elements from a pool of random sequences. Mol. Cell. Biol. 15, 6291–6298 (1995).
Cartegni, L., Wang, J., Zhu, Z., Zhang, M. Q. & Krainer, A. R. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 31, 3568–3571 (2003).
Coulter, L. R., Landree, M. A. & Cooper, T. A. Identification of a new class of exonic splicing enhancers by in vivo selection. Mol. Cell. Biol. 17, 2143–2150 (1997). A functional SELEX approach, based on the recovery of sequences that led to exon inclusion, allowed the identification of various ESEs, including a novel AC-rich class.
Stickeler, E. et al. The RNA binding protein YB-1 binds A/C-rich exon enhancers and stimulates splicing of the CD44 alternative exon v4. EMBO J. 20, 3821–3830 (2001).
Fedorov, A., Saxonov, S., Fedorova, L. & Daizadeh, I. Comparison of intron-containing and intron-lacking human genes elucidates putative exonic splicing enhancers. Nucleic Acids Res. 29, 1464–1469 (2001).
Fairbrother, W. G., Yeh, R. F., Sharp, P. A. & Burge, C. B. Predictive identification of exonic splicing enhancers in human genes. Science 297, 1007–1013 (2002). A computational survey for ESEs, based on the enrichment in exons versus introns, and in exons with weak splice sites versus exons with strong splice sites; led to the development of the RESCUE-ESE web resource.
Yeo, G., Hoon, S., Venkatesh, B. & Burge, C. B. Variation in sequence and organization of splicing regulatory elements in vertebrate genes. Proc. Natl Acad. Sci. USA 101, 15700–15705 (2004).
Minovitsky, S., Gee, S. L., Schokrpur, S., Dubchak, I. & Conboy, J. G. The splicing regulatory element, UGCAUG, is phylogenetically and spatially conserved in introns that flank tissue-specific alternative exons. Nucleic Acids Res. 33, 714–724 (2005).
Miriami, E., Margalit, H. & Sperling, R. Conserved sequence elements associated with exon skipping. Nucleic Acids Res. 31, 1974–1983 (2003).
Yeo, G., Holste, D., Kreiman, G. & Burge, C. B. Variation in alternative splicing across human tissues. Genome Biol. 5, R74 (2004). Analysis of publicly available tissue-specific EST and microarray expression data showed that liver had the most distinctive patterns of alternative splicing and expression of SR and hnRNP proteins.
Tenenbaum, S. A., Carson, C. C., Lager, P. J. & Keene, J. D. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc. Natl Acad. Sci. USA 97, 14085–14090 (2000).
Blanchette, M., Labourier, E., Green, R. E., Brenner, S. E. & Rio, D. C. Genome-wide analysis reveals an unexpected function for the Drosophila splicing factor U2AF(50) in the nuclear export of intronless mRNAs. Mol. Cell 14, 775–786 (2004). Conditional mutants for the D. melanogaster large subunit of U2AF and RNAi knockdown indicate that the U2AF large subunit has an unexpected role in nuclear export of intronless RNAs.
Mili, S. & Steitz, J. A. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA 10, 1692–1694 (2004).
Buckanovich, R. J. & Darnell, R. B. The neuronal RNA binding protein Nova-1 recognizes specific RNA targets in vitro and in vivo. Mol. Cell. Biol. 17, 3194–3201 (1997).
Kim, S., Shi, H., Lee, D. K. & Lis, J. T. Specific SR protein-dependent splicing substrates identified through genomic SELEX. Nucleic Acids Res. 31, 1955–1961 (2003). Successful application of genomic SELEX to identify cellular RNA targets of D. melanogaster B52 (SRp55), a number of which were validated in functional assays.
Hieronymous, H. & Silver, P. A. A systems view of mRNP biology. Genes Dev. 18, 2845–2860 (2004).
Kotovic, K. M., Lockshon, D., Boric, L. & Neugebauer, K. M. Cotranscriptional recruitment of the U1 snRNP to intron-containing genes in yeast. Mol. Cell. Biol. 23, 5768–5779 (2003).
Longman, D., Johnstone, I. L. & Caceres, J. F. Functional characterization of SR and SR-related genes in Caenorhabditis elegans. EMBO J. 19, 1625–1637 (2000).
Wang, H. Y., Xu, X., Ding, J. H., Bermingham, J. R. Jr & Fu, X. D. SC35 plays a role in T cell development and alternative splicing of CD45. Mol. Cell 7, 331–342 (2001).
Huang, Y. & Steitz, J. A. SRprises along a messenger's journey. Mol. Cell 17, 613–615 (2005).
Rélogio, A. et al. Alternative splicing microarrays reveal functional expression of neuron specific regulators in Hodgkin lymphoma cells. J. Biol. Chem. 280, 4779–4784 (2005). Used a microarray that contained probes specific for alternative splicing events as well as the expression of splicing regulators. This design potentially allows the deciphering of splicing factor codes that regulate particular splicing events.
Pan, Q. et al. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol. Cell 16, 929–941 (2004). A quantitative alternative splicing microarray analysed 3,126 known alternative splicing events, allowing many new insights into the global aspects of alternative splicing programmes.
Wollerton, M. C., Gooding, C., Wagner, E. J., Garcia-Blanco, M. A. & Smith, C. W. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol. Cell 13, 91–100 (2004). Demonstrates a negative feedback loop in which PTB promotes skipping of its own exon 11, producing an RNA that is degraded by NMD.
Park, J. W., Parisky, K., Celotto, A. M., Reenan, R. A. & Graveley, B. R. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc. Natl Acad. Sci. USA 101, 15974–15979 (2004). An RNAi screen allowed the identification of a large number of splicing regulators, including a number of constitutive splicing factors.
Lim, S. R. & Hertel, K. J. Commitment to splice site pairing coincides with A complex formation. Mol. Cell 15, 477–483 (2004). Shows that although pre-mRNAs are committed to splicing in the E complex, the decision as to which splice sites will be paired is subsequently made in the A complex.
Berget, S. M. Exon recognition in vertebrate splicing. J. Biol. Chem. 270, 2411–2414 (1995).
Chiara, M. D. & Reed, R. A two-step mechanism for 5′ and 3′ splice-site pairing. Nature 375, 510–513 (1995).
Niranjanakumari, S., Lasda, E., Brazas, R. & Garcia-Blanco, M. A. Reversible crosslinking combined with immunoprecipitation to study RNA–protein interactions in vivo. Methods 26, 182–190 (2002).
Wang, H. et al. Gene structure-based splice variant deconvolution using a microarray platform. Bioinformatics 19 (Suppl. 1), i315–i322 (2003).
Hiller, M. et al. Widespread occurrence of alternative splicing at NAGNAG acceptors contributes to proteome plasticity. Nature Genet. 36, 1255–1257 (2004).
Acknowledgements
We thank Juan Valcárcel and Sushma Nagaraja Grellscheid for critically reading the manuscript. Work in the laboratory of C.W.J.S. is supported by a programme grant from the Wellcome Trust. We apologize to those colleagues whose work, for reasons of space constraints, we have been unable to cite directly.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- ENHANCER
-
A cis-acting RNA sequence in an exon (ESE) or intron (ISE) on which a complex, often containing SR proteins, assembles to promote the use of a weak or regulated splice site.
- SILENCER
-
A cis-acting RNA sequence in an exon (ESS) or intron (ISS) on which a complex, often containing heterogeneous nuclear ribonucleoproteins, assembles to repress the use of a splice site.
- 'GENOME TILING' MICROARRAY
-
A type of microarray in which overlapping oligonucleotides are designed to cover a genomic region of interest.
- NONSENSE-MEDIATED DECAY
-
(NMD). A surveillance pathway leading to the targeted degradation of mRNAs that contain premature termination codons. In mammals, termination codons that lie more than 50–55 nucleotides upstream of an exon–exon junction are recognized as premature.
- EXON DEFINITION
-
The initial interaction and stabilization across an exon of factors bound to the 3′ and 5′ splice sites prior to the formation of cross-intron bridging interactions.
- SR FAMILY OF PROTEINS
-
Required for both constitutive and regulated splicing, with a modular structure comprising one or two N-terminal RRMs and a C-terminal arginine/serine-rich (RS) domain.
- RNA RECOGNITION MOTIF
-
(RMM). A protein domain that is frequently involved in sequence-specific single-stranded RNA binding, and consists of a βαββαβ fold with the β strands forming a surface that displays two highly conserved RNP motifs and forms contacts with the RNA.
- HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN
-
(hnRNP). A group of >20 proteins that associate with high-molecular-weight nuclear RNA. Some hnRNPs, such as members of the hnRNPA/B family, shuttle in and out of the nucleus, whereas others are strictly nuclear.
- KH
-
An hnRNPK homology domain that binds to single-stranded RNA, and contains α and β secondary structure elements.
- SMALL NUCLEAR RIBONUCLEOPROTEIN PARTICLE
-
(snRNP). snRNPs U1, U2, U4, U5 and U6, which contain proteins and U-rich RNA, are core components of the spliceosome.
- ARTIFICIAL RECRUITMENT
-
An experimental approach that allows the analysis of 'effector' domains (repressors or activators), independent of the ability to bind RNA, by fusing an RNA-binding domain (typically, the bacteriophage MS2 coat protein) to the protein or domain of interest, and by introducing cognate RNA-binding sites into the reporter RNA.
- BRANCH POINT
-
An adenosine within a variably conserved 'branch point sequence' upstream of the 3′ splice site that is involved in the first chemical step of splicing. A new 2′–5′ phosphodiester is formed between the branch point and the 5′ end of the intron, concomitant with breakage of the link between the 5′ exon and the intron.
- PARALOGUE
-
A sequence, or gene, that originates from a common ancestral sequence, or gene, by a duplication event.
- SELEX
-
(Systematic evolution of ligands by exponential enrichment). An iterative method for the selection of optimal sites for RNA-binding proteins. Starting from a pool of degenerate cDNAs, sequences are subjected to several rounds of transcription, protein binding and RT-PCR until a consensus sequence emerges.
- PRE-mRNP
-
(or mRNP). A ribonucleoprotein complex, of variable protein composition, that is associated with pre-mRNAs or mRNAs, respectively.
Rights and permissions
About this article
Cite this article
Matlin, A., Clark, F. & Smith, C. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol 6, 386–398 (2005). https://doi.org/10.1038/nrm1645
Issue Date:
DOI: https://doi.org/10.1038/nrm1645
This article is cited by
-
Estimating transcriptome complexities across eukaryotes
BMC Genomics (2023)
-
SEPepQuant enhances the detection of possible isoform regulations in shotgun proteomics
Nature Communications (2023)
-
Context-aware transcript quantification from long-read RNA-seq data with Bambu
Nature Methods (2023)
-
Investigating open reading frames in known and novel transcripts using ORFanage
Nature Computational Science (2023)
-
The implications of alternative pre-mRNA splicing in cell signal transduction
Experimental & Molecular Medicine (2023)