Abstract
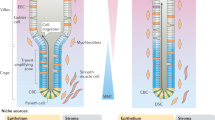
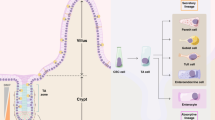
The intestinal epithelium withstands continuous mechanical, chemical and biological insults despite its single-layered, simple epithelial structure. The crypt–villus tissue architecture in combination with rapid cell turnover enables the intestine to act both as a barrier and as the primary site of nutrient uptake. Constant tissue replenishment is fuelled by continuously dividing stem cells that reside at the bottom of crypts. These cells are nurtured and protected by specialized epithelial and mesenchymal cells, and together constitute the intestinal stem cell niche. Intestinal stem cells and early progenitor cells compete for limited niche space and, therefore, the ability to retain or regain stemness. Those cells unable to do so differentiate to one of six different mature cell types and move upwards towards the villus, where they are shed into the intestinal lumen after 3–5 days. In this Review, we discuss the signals, cell types and mechanisms that control homeostasis and regeneration in the intestinal epithelium. We investigate how the niche protects and instructs intestinal stem cells, which processes drive differentiation of mature cells and how imbalance in key signalling pathways can cause human disease.
Key points
-
Crypt–villus structure and continuous proliferation enable the intestine to act as an absorptive organ and a protective barrier.
-
Tissue replenishment is fuelled by adult stem cells that divide continuously and reside sequestered away from the intestinal lumen at the bottom of crypts.
-
Stem cells are protected by their niche (Paneth cells and the surrounding mesenchyme) through crypt shape, antimicrobial products, a specialized metabolic environment and neutral competition for limited space.
-
Stemness is a state that is lost when cells leave the stem cell zone, but can also be regained when differentiated cells re-enter the niche.
-
Together with interleukins, Hippo signalling and metabolic cues, WNT, NOTCH, EGF and bone morphogenetic protein signalling regulates stem cell maintenance, regeneration and differentiation.
-
The same signalling cascades involved in stem cell maintenance are key regulators of intestinal differentiation and cooperate to guide cells to one of six mature cell fates.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Helander, H. F. & Fandriks, L. Surface area of the digestive tract — revisited. Scand. J. Gastroenterol. 49, 681–689 (2014).
Frohlich, E., Mercuri, A., Wu, S. & Salar-Behzadi, S. Measurements of deposition, lung surface area and lung fluid for simulation of inhaled compounds. Front. Pharmacol. 7, 181 (2016).
Lieberkuehn, J. N. Dissertatio Anatomico Physiologica de Fabrica et Actione Villorum Intestinorum Tenuium Hominis (apud Conrad et Georg. Jac. Wishof, 1745).
Darwich, A. S., Aslam, U., Ashcroft, D. M. & Rostami-Hodjegan, A. Meta-analysis of the turnover of intestinal epithelia in preclinical animal species and humans. Drug Metab. Dispos. 42, 2016–2022 (2014).
Cheng, H. & Leblond, C. P. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. V. Unitarian gheory of the origin of the four epithelial cell types. Am. J. Anat. 141, 537–561 (1974).
Winton, D. J., Blount, M. A. & Ponder, B. A. A clonal marker induced by mutation in mouse intestinal epithelium. Nature 333, 463–466 (1988).
Bjerknes, M. & Cheng, H. Clonal analysis of mouse intestinal epithelial progenitors. Gastroenterology 116, 7–14 (1999).
van de Wetering, M. et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 111, 241–250 (2002).
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007).This paper describes the discovery of Lgr5 as marker of adult stem cells in the intestine.
Potten, C. S., Hume, W. J., Reid, P. & Cairns, J. The segregation of DNA in epithelial stem cells. Cell 15, 899–906 (1978).
Potten, C. S., Owen, G. & Booth, D. Intestinal stem cells protect their genome by selective segregation of template DNA strands. J. Cell Sci. 115, 2381–2388 (2002).
Sangiorgi, E. & Capecchi, M. R. Bmi1 is expressed in vivo in intestinal stem cells. Nat. Genet. 40, 915–920 (2008).
Breault, D. T. et al. Generation of mTert-GFP mice as a model to identify and study tissue progenitor cells. Proc. Natl Acad. Sci. USA 105, 10420–10425 (2008).
Montgomery, R. K. et al. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc. Natl Acad. Sci. USA 108, 179–184 (2011).
Takeda, N. et al. Interconversion between intestinal stem cell populations in distinct niches. Science 334, 1420–1424 (2011).
Powell, A. E. et al. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell 149, 146–158 (2012).
Wong, V. W. et al. Lrig1 controls intestinal stem-cell homeostasis by negative regulation of ErbB signalling. Nat. Cell Biol. 14, 401–408 (2012).
Grun, D. et al. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature 525, 251–255 (2015).
Munoz, J. et al. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. EMBO J. 31, 3079–3091 (2012).
Wang, F. et al. Isolation and characterization of intestinal stem cells based on surface marker combinations and colony-formation assay. Gastroenterology 145, 383–395.e21 (2013).
Li, N. et al. Single-cell analysis of proxy reporter allele-marked epithelial cells establishes intestinal stem cell hierarchy. Stem Cell Rep. 3, 876–891 (2014).
Li, N., Nakauka-Ddamba, A., Tobias, J., Jensen, S. T. & Lengner, C. J. Mouse label-retaining cells are molecularly and functionally distinct from reserve intestinal stem cells. Gastroenterology 151, 298–310.e7 (2016).
Buczacki, S. J. et al. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature 495, 65–69 (2013).This elegant study marked, lineage-traced and identified label-retaining cells in the intestine.
Roche, K. C. et al. SOX9 maintains reserve stem cells and preserves radioresistance in mouse small intestine. Gastroenterology 149, 1553–1563.e10 (2015).
Tetteh, P. W., Farin, H. F. & Clevers, H. Plasticity within stem cell hierarchies in mammalian epithelia. Trends Cell Biol. 25, 100–108 (2015).
Dekaney, C. M., Gulati, A. S., Garrison, A. P., Helmrath, M. A. & Henning, S. J. Regeneration of intestinal stem/progenitor cells following doxorubicin treatment of mice. Am. J. Physiol. Gastrointest. Liver Phsiol. 297, G461–G470 (2009).
van Es, J. H. et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat. Cell Biol. 14, 1099–1104 (2012).This study was the first report of secretory progenitors being able to regain stemness upon intestinal tissue damage.
Tian, H. et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature 478, 255–259 (2011).
Metcalfe, C., Kljavin, N. M., Ybarra, R. & de Sauvage, F. J. Lgr5+ stem cells are indispensable for radiation-induced intestinal regeneration. Cell Stem Cell 14, 149–159 (2014).
Yan, K. S. et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc. Natl Acad. Sci. USA 109, 466–471 (2012).
Yan, K. S. et al. Intestinal enteroendocrine lineage cells possess homeostatic and injury-inducible stem cell activity. Cell Stem Cell 21, 78–90.e6 (2017).
Jadhav, U. et al. Dynamic reorganization of chromatin accessibility signatures during dedifferentiation of secretory precursors into Lgr5+ intestinal stem cells. Cell Stem Cell 21, 65–77.e5 (2017).
Tetteh, P. W. et al. Replacement of lost Lgr5-positive stem cells through plasticity of their enterocyte-lineage daughters. Cell Stem Cell 18, 203–213 (2016). This study was the first report that intestinal plasticity is not limited to the secretory lineage but can also be found in absorptive cells.
Kaaij, L. T. et al. DNA methylation dynamics during intestinal stem cell differentiation reveals enhancers driving gene expression in the villus. Genome Biol. 14, R50 (2013).
Yu, D. H. et al. Postnatal epigenetic regulation of intestinal stem cells requires DNA methylation and is guided by the microbiome. Genome Biol. 16, 211 (2015).
Kim, T. H. et al. Broadly permissive intestinal chromatin underlies lateral inhibition and cell plasticity. Nature 506, 511–515 (2014).
Jadhav, U. et al. Acquired tissue-specific promoter bivalency is a basis for PRC2 necessity in adult cells. Cell 165, 1389–1400 (2016).
Tata, P. R. et al. Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature 503, 218–223 (2013).
Stange, D. E. et al. Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell 155, 357–368 (2013).
Tarlow, B. D. et al. Bipotential adult liver progenitors are derived from chronically injured mature hepatocytes. Cell Stem Cell 15, 605–618 (2014).
Kaiko, G. E. et al. The colonic crypt protects stem cells from microbiota-derived metabolites. Cell 167, 1137 (2016).
Liu, J., Walker, N. M., Cook, M. T., Ootani, A. & Clarke, L. L. Functional Cftr in crypt epithelium of organotypic enteroid cultures from murine small intestine. Am. J. Physiol. Cell Physiol. 302, C1492–C1503 (2012).
Trezise, A. E. et al. The multidrug resistance and cystic fibrosis genes have complementary patterns of epithelial expression. EMBO J. 11, 4291–4303 (1992).
Johansson, M. E., Larsson, J. M. & Hansson, G. C. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc. Natl Acad. Sci. USA 108, S4659–S4665 (2011).
Birchenough, G. M., Nystrom, E. E., Johansson, M. E. & Hansson, G. C. A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science 352, 1535–1542 (2016).
Batlle, E. et al. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell 111, 251–263 (2002).
Genander, M. et al. Dissociation of EphB2 signaling pathways mediating progenitor cell proliferation and tumor suppression. Cell 139, 679–692 (2009).
Gassler, N. Paneth cells in intestinal physiology and pathophysiology. World J. Gastrointest. Pathophysiol. 8, 150–160 (2017).
Allaire, J. M. et al. The intestinal epithelium: central coordinator of mucosal immunity. Trends Immunol. 39, 677–696 (2018).
Sato, T. et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469, 415–418 (2011).This study was the first report of Paneth cells constituting the epithelial niche in the small intestine.
Sasaki, N. et al. Reg4+ deep crypt secretory cells function as epithelial niche for Lgr5+ stem cells in colon. Proc. Natl Acad. Sci. USA 113, E5399–E5407 (2016).
Rodriguez-Colman, M. J. et al. Interplay between metabolic identities in the intestinal crypt supports stem cell function. Nature 543, 424–427 (2017).This study characterized the metabolic features of CBC cells and Paneth cells in the intestinal crypt and established reactive oxygen species as drivers for differentiation.
Blokzijl, F. et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature 538, 260–264 (2016).
Groussard, C. et al. Free radical scavenging and antioxidant effects of lactate ion: an in vitro study. J. Appl. Physiol. 89, 169–175 (2000).
Snippert, H. J. et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144 (2010).
Lopez-Garcia, C., Klein, A. M., Simons, B. D. & Winton, D. J. Intestinal stem cell replacement follows a pattern of neutral drift. Science 330, 822–825 (2010).References 55 and 56 were the initial publications describing neutral competition of intestinal stem cells in the niche.
Vermeulen, L. et al. Defining stem cell dynamics in models of intestinal tumor initiation. Science 342, 995–998 (2013).
Snippert, H. J., Schepers, A. G., van Es, J. H., Simons, B. D. & Clevers, H. Biased competition between Lgr5 intestinal stem cells driven by oncogenic mutation induces clonal expansion. EMBO Rep. 15, 62–69 (2014).
Cancer Genome Atlas, N. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012).
Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009).This study is the first description of intestinal organoid culture.
Langlands, A. J. et al. Paneth cell-rich regions separated by a cluster of Lgr5+ cells initiate crypt fission in the intestinal stem cell niche. PLOS Biol. 14, e1002491 (2016).
Ritsma, L. et al. Intestinal crypt homeostasis revealed at single-stem-cell level by in vivo live imaging. Nature 507, 362–365 (2014).
Clevers, H. C. & Bevins, C. L. Paneth cells: maestros of the small intestinal crypts. Annu. Rev. Physiol. 75, 289–311 (2013).
Durand, A. et al. Functional intestinal stem cells after Paneth cell ablation induced by the loss of transcription factor Math1 (Atoh1). Proc. Natl Acad. Sci. USA 109, 8965–8970 (2012).
Kim, T. H., Escudero, S. & Shivdasani, R. A. Intact function of Lgr5 receptor-expressing intestinal stem cells in the absence of Paneth cells. Proc. Natl Acad. Sci. USA 109, 3932–3937 (2012).
Sancho, R., Cremona, C. A. & Behrens, A. Stem cell and progenitor fate in the mammalian intestine: Notch and lateral inhibition in homeostasis and disease. EMBO Rep. 16, 571–581 (2015).
Bastide, P. et al. Sox9 regulates cell proliferation and is required for Paneth cell differentiation in the intestinal epithelium. J. Cell Biol. 178, 635–648 (2007).
Mori-Akiyama, Y. et al. SOX9 is required for the differentiation of paneth cells in the intestinal epithelium. Gastroenterology 133, 539–546 (2007).
Shroyer, N. F., Wallis, D., Venken, K. J., Bellen, H. J. & Zoghbi, H. Y. Gfi1 functions downstream of Math1 to control intestinal secretory cell subtype allocation and differentiation. Genes Dev. 19, 2412–2417 (2005).
Aoki, R. et al. Foxl1-expressing mesenchymal cells constitute the intestinal stem cell niche. Cell. Mol. Gastroenterol. Hepatol. 2, 175–188 (2016).
Valenta, T. et al. Wnt ligands secreted by subepithelial mesenchymal cells are essential for the survival of intestinal stem cells and gut homeostasis. Cell Rep. 15, 911–918 (2016).
Stzepourginski, I. et al. CD34+ mesenchymal cells are a major component of the intestinal stem cells niche at homeostasis and after injury. Proc. Natl Acad. Sci. USA 114, E506–E513 (2017).
Mao, J., Kim, B. M., Rajurkar, M., Shivdasani, R. A. & McMahon, A. P. Hedgehog signaling controls mesenchymal growth in the developing mammalian digestive tract. Development 137, 1721–1729 (2010).
Zacharias, W. J. et al. Hedgehog signaling controls homeostasis of adult intestinal smooth muscle. Dev. Biol. 355, 152–162 (2011).
Nusse, R. & Clevers, H. Wnt/beta-catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985–999 (2017).
Korinek, V. et al. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat. Genet. 19, 379–383 (1998).
van Es, J. H. et al. A critical role for the Wnt effector Tcf4 in adult intestinal homeostatic self-renewal. Mol. Cell. Biol. 32, 1918–1927 (2012).
Kuhnert, F. et al. Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proc. Natl Acad. Sci. USA 101, 266–271 (2004).
Pinto, D., Gregorieff, A., Begthel, H. & Clevers, H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 17, 1709–1713 (2003).
Barker, N. et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 457, 608–611 (2009).
Moser, A. R. et al. ApcMin: a mouse model for intestinal and mammary tumorigenesis. Eur. J. Cancer 31, 1061–1064 (1995).
Alexandre, C., Baena-Lopez, A. & Vincent, J. P. Patterning and growth control by membrane-tethered Wingless. Nature 505, 180–185 (2014).
Farin, H. F. et al. Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature 530, 340–343 (2016).
Farin, H. F., Van Es,J. H. & Clevers, H. Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology 143, 1518–1529.e7 (2012).
de Lau, W. et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature 476, 293–297 (2011).
Glinka, A. et al. LGR4 and LGR5 are R-spondin receptors mediating Wnt/beta-catenin and Wnt/PCP signalling. EMBO Rep. 12, 1055–1061 (2011).
Carmon, K. S., Gong, X., Lin, Q., Thomas, A. & Liu, Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc. Natl Acad. Sci. USA 108, 11452–11457 (2011).
Koo, B. K. et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature 488, 665–669 (2012).
Hao, H. X. et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature 485, 195–200 (2012).
de Lau, W., Peng, W. C., Gros, P. & Clevers, H. The R-spondin/Lgr5/Rnf43 module: regulator of Wnt signal strength. Genes Dev. 28, 305–316 (2014).
Storm, E. E. et al. Targeting PTPRK-RSPO3 colon tumours promotes differentiation and loss of stem-cell function. Nature 529, 97–100 (2016).
van Landeghem, L. et al. Enteric glia promote intestinal mucosal healing via activation of focal adhesion kinase and release of proEGF. Am. J. Physiol. Gastrointest. Liver Phsiol. 300, G976–G987 (2011).
Basak, O. et al. Induced quiescence of Lgr5+ stem cells in intestinal organoids enables differentiation of hormone-producing enteroendocrine cells. Cell Stem Cell 20, 177–190.e4 (2017).
Tian, H. et al. Opposing activities of Notch and Wnt signaling regulate intestinal stem cells and gut homeostasis. Cell Rep. 11, 33–42 (2015).
van Es, J. H. et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature 435, 959–963 (2005).
Milano, J. et al. Modulation of notch processing by gamma-secretase inhibitors causes intestinal goblet cell metaplasia and induction of genes known to specify gut secretory lineage differentiation. Toxicol. Sci. 82, 341–358 (2004).
VanDussen, K. L. & Samuelson, L. C. Mouse atonal homolog 1 directs intestinal progenitors to secretory cell rather than absorptive cell fate. Dev. Biol. 346, 215–223 (2010).
Shroyer, N. F. et al. Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology 132, 2478–2488 (2007).
Li, H. J., Kapoor, A., Giel-Moloney, M., Rindi, G. & Leiter, A. B. Notch signaling differentially regulates the cell fate of early endocrine precursor cells and their maturing descendants in the mouse pancreas and intestine. Dev. Biol. 371, 156–169 (2012).
Takebayashi, K. et al. Structure, chromosomal locus, and promoter analysis of the gene encoding the mouse helix-loop-helix factor HES-1: negative autoregulation through the multiple N box elements. J. Biol. Chem. 269, 5150–5156 (1994).
Hirata, H. et al. Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science 298, 840–843 (2002).
Yamamizu, K. et al. Convergence of Notch and beta-catenin signaling induces arterial fate in vascular progenitors. J. Cell Biol. 189, 325–338 (2010).
Kay, S. K. et al. The role of the Hes1 crosstalk hub in Notch-Wnt interactions of the intestinal crypt. PLOS Computat. Biol. 13, e1005400 (2017).
Massague, J. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 13, 616–630 (2012).
Hardwick, J. C. et al. Bone morphogenetic protein 2 is expressed by, and acts upon, mature epithelial cells in the colon. Gastroenterology 126, 111–121 (2004).
Haramis, A. P. et al. De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science 303, 1684–1686 (2004).
He, X. C. et al. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-beta-catenin signaling. Nat. Genet. 36, 1117–1121 (2004).
Kosinski, C. et al. Gene expression patterns of human colon tops and basal crypts and BMP antagonists as intestinal stem cell niche factors. Proc. Natl Acad. Sci. USA 104, 15418–15423 (2007).
Davis, H. et al. Aberrant epithelial GREM1 expression initiates colonic tumorigenesis from cells outside the stem cell niche. Nat. Med. 21, 62–70 (2015).
Ma, H. et al. Pathology and genetics of hereditary colorectal cancer. Pathology 50, 49–59 (2018).
Qi, Z. et al. BMP restricts stemness of intestinal Lgr5+ stem cells by directly suppressing their signature genes. Nat. Commun. 8, 13824 (2017).
Jiang, H. et al. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell 137, 1343–1355 (2009).
Hanash, A. M. et al. Interleukin-22 protects intestinal stem cells from immune-mediated tissue damage and regulates sensitivity to graft versus host disease. Immunity 37, 339–350 (2012).
Zenewicz, L. A. et al. Interleukin-22 but not interleukin-17 provides protection to hepatocytes during acute liver inflammation. Immunity 27, 647–659 (2007).
Sugimoto, K. et al. IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J. Clin. Invest. 118, 534–544 (2008).
Lindemans, C. A. et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature 528, 560–564 (2015).
Gerbe, F. et al. Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature 529, 226–230 (2016).
von Moltke, J., Ji, M., Liang, H. E. & Locksley, R. M. Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature 529, 221–225 (2016).References 117 and 118 clarified the function of Tuft cells in the intestine.
Meng, Z., Moroishi, T. & Guan, K. L. Mechanisms of Hippo pathway regulation. Genes Dev. 30, 1–17 (2016).
Gregorieff, A. & Wrana, J. L. Hippo signalling in intestinal regeneration and cancer. Curr. Opin. Cell Biol. 48, 17–25 (2017).
Barry, E. R. et al. Restriction of intestinal stem cell expansion and the regenerative response by YAP. Nature 493, 106–110 (2013).
Cai, J. et al. The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes Dev. 24, 2383–2388 (2010).
Gregorieff, A., Liu, Y., Inanlou, M. R., Khomchuk, Y. & Wrana, J. L. Yap-dependent reprogramming of Lgr5(+) stem cells drives intestinal regeneration and cancer. Nature 526, 715–718 (2015).
Imajo, M., Ebisuya, M. & Nishida, E. Dual role of YAP and TAZ in renewal of the intestinal epithelium. Nat. Cell Biol. 17, 7–19 (2015).
Zhou, D. et al. Mst1 and Mst2 protein kinases restrain intestinal stem cell proliferation and colonic tumorigenesis by inhibition of Yes-associated protein (Yap) overabundance. Proc. Natl Acad. Sci. USA 108, E1312–E1320 (2011).
Varelas, X. et al. The Hippo pathway regulates Wnt/beta-catenin signaling. Dev. Cell 18, 579–591 (2010).
Azzolin, L. et al. YAP/TAZ incorporation in the beta-catenin destruction complex orchestrates the Wnt response. Cell 158, 157–170 (2014).
Park, H. W. et al. Alternative Wnt signaling activates YAP/TAZ. Cell 162, 780–794 (2015).
Andrew, A. L. et al. Rapid changes in gene expression direct rapid shifts in intestinal form and function in the Burmese python after feeding. Physiol. Genomics 47, 147–157 (2015).
Dunel-Erb, S. et al. Restoration of the jejunal mucosa in rats refed after prolonged fasting. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 129, 933–947 (2001).
Galluser, M. et al. Adaptation of intestinal hydrolases to starvation in rats: effect of thyroid function. J. Comp. Physiol. B 161, 357–361 (1991).
Richmond, C. A. et al. Dormant intestinal stem cells are regulated by PTEN and nutritional status. Cell Rep. 13, 2403–2411 (2015).
Yilmaz, O. H. et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature 486, 490–495 (2012).
Tinkum, K. L. et al. Fasting protects mice from lethal DNA damage by promoting small intestinal epithelial stem cell survival. Proc. Natl Acad. Sci. USA 112, E7148–E7154 (2015).
Beyaz, S. et al. High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature 531, 53–58 (2016).
Bardou, M., Barkun, A. N. & Martel, M. Obesity and colorectal cancer. Gut 62, 933–947 (2013).
van Es, J. H. et al. Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nat. Cell Biol. 7, 381–386 (2005).
Vidrich, A. et al. Fibroblast growth factor receptor-3 regulates Paneth cell lineage allocation and accrual of epithelial stem cells during murine intestinal development. Am. J. Physiol. Gastrointest. Liver Physiol. 297, G168–G178 (2009).
Heuberger, J. et al. Shp2/MAPK signaling controls goblet/paneth cell fate decisions in the intestine. Proc. Natl Acad. Sci. USA 111, 3472–3477 (2014).
Gregorieff, A. et al. The ets-domain transcription factor Spdef promotes maturation of goblet and paneth cells in the intestinal epithelium. Gastroenterology 137, 1333–1345.e3 (2009).
Noah, T. K., Kazanjian, A., Whitsett, J. & Shroyer, N. F. SAM pointed domain ETS factor (SPDEF) regulates terminal differentiation and maturation of intestinal goblet cells. Exp. Cell Res. 316, 452–465 (2010).
Worthington, J. J., Reimann, F. & Gribble, F. M. Enteroendocrine cells-sensory sentinels of the intestinal environment and orchestrators of mucosal immunity. Mucosal Immunol. 11, 3–20 (2018).
Lopez-Diaz, L. et al. Intestinal Neurogenin 3 directs differentiation of a bipotential secretory progenitor to endocrine cell rather than goblet cell fate. Dev. Biol. 309, 298–305 (2007).
Jenny, M. et al. Neurogenin3 is differentially required for endocrine cell fate specification in the intestinal and gastric epithelium. EMBO J. 21, 6338–6347 (2002).
Gerbe, F., Legraverend, C. & Jay, P. The intestinal epithelium tuft cells: specification and function. Cell. Mol. Life Sci. 69, 2907–2917 (2012).
Bjerknes, M. et al. Origin of the brush cell lineage in the mouse intestinal epithelium. Dev. Biol. 362, 194–218 (2012).
Gerbe, F. et al. Distinct ATOH1 and Neurog3 requirements define tuft cells as a new secretory cell type in the intestinal epithelium. J. Cell Biol. 192, 767–780 (2011).
Ross, M. H. & Pawlina, W. Histology: A Text and Atlas: with Correlated Cell and Molecular Biology 5th edn (Lippincott Wiliams & Wilkins, 2006).
Yang, Q., Bermingham, N. A., Finegold, M. J. & Zoghbi, H. Y. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science 294, 2155–2158 (2001).
Mabbott, N. A., Donaldson, D. S., Ohno, H., Williams, I. R. & Mahajan, A. Microfold (M) cells: important immunosurveillance posts in the intestinal epithelium. Mucosal Immunol. 6, 666–677 (2013).
Knoop, K. A. et al. RANKL is necessary and sufficient to initiate development of antigen-sampling M cells in the intestinal epithelium. J. Immunol. 183, 5738–5747 (2009).
de Lau, W. et al. Peyer’s patch M cells derived from Lgr5(+) stem cells require SpiB and are induced by RankL in cultured “miniguts”. Mol. Cell. Biol. 32, 3639–3647 (2012).
Kanaya, T. et al. The Ets transcription factor Spi-B is essential for the differentiation of intestinal microfold cells. Nat. Immunol. 13, 729–736 (2012).
Laurenti, E. & Gottgens, B. From haematopoietic stem cells to complex differentiation landscapes. Nature 553, 418–426 (2018).
van de Wetering, M. et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945 (2015).
Drost, J. et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 521, 43–47 (2015). This study elegantly used stepwise-directed mutagenesis to transform healthy human intestinal epithelium into a carcinoma.
Fumagalli, A. et al. Genetic dissection of colorectal cancer progression by orthotopic transplantation of engineered cancer organoids. Proc. Natl Acad. Sci. USA 114, E2357–E2364 (2017).
Vlachogiannis, G. et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 359, 920–926 (2018).
Dekkers, J. F. et al. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat. Med. 19, 939–945 (2013).
Dekkers, J. F. et al. Characterizing responses to CFTR-modulating drugs using rectal organoids derived from subjects with cystic fibrosis. Sci. Transl Med. 8, 344ra84 (2016).
Yui, S. et al. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5(+) stem cell. Nat. Med. 18, 618–623 (2012).
Fukuda, M. et al. Small intestinal stem cell identity is maintained with functional Paneth cells in heterotopically grafted epithelium onto the colon. Genes Dev. 28, 1752–1757 (2014).
Cruz-Acuna, R. et al. Synthetic hydrogels for human intestinal organoid generation and colonic wound repair. Nat. Cell Biol. 19, 1326–1335 (2017).
Sugimoto, S. et al. Reconstruction of the human colon epithelium in vivo. Cell Stem Cell 22, 171–176.e5 (2017). This study is the first report of successful transplantation of human intestinal organoids in mouse intestine.
Schwank, G. et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 13, 653–658 (2013).
Acknowledgements
The authors thank J. van Es for critical reading of the manuscript. H.G. was supported by VENI fellowship 016.16.119 of the Netherlands Organisation for Scientific Research NWO.
Reviewer information
Nature Reviews Gastroenterology & Hepatology thanks K. Yan and the other anonymous reviewers(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
H.G. researched data for the article and wrote the manuscript. H.G. and H.C. contributed equally to discussion of content and reviewing/editing the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Gehart, H., Clevers, H. Tales from the crypt: new insights into intestinal stem cells. Nat Rev Gastroenterol Hepatol 16, 19–34 (2019). https://doi.org/10.1038/s41575-018-0081-y
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41575-018-0081-y
This article is cited by
-
Ginseng-derived nanoparticles alleviate inflammatory bowel disease via the TLR4/MAPK and p62/Nrf2/Keap1 pathways
Journal of Nanobiotechnology (2024)
-
The gut microbiota and its biogeography
Nature Reviews Microbiology (2024)
-
SLE serum induces altered goblet cell differentiation and leakiness in human intestinal organoids
EMBO Molecular Medicine (2024)
-
Drosophila activins adapt gut size to food intake and promote regenerative growth
Nature Communications (2024)
-
Distal colonocytes targeted by C. rodentium recruit T-cell help for barrier defence
Nature (2024)