Abstract
We investigated the quantitative prediction of human hepatic metabolic clearance from in vitro experiments focusing on cytochrome P450 metabolism with eight model compounds, FK1052, FK480, zolpidem, omeprazole, nicardipine, nilvadipine, diazepam, and diltiazem. For the compounds, in vivo human hepatic extraction ratios ranged widely from 0.03 to 0.87. In vitro and in vivo hepatic intrinsic clearance (CLint) values for each compound were measured and calculated in rats and/or dogs and humans. CLint,in vitro was determined from a substrate disappearance rate at 1 μM in hepatic microsomes, which was a useful method. CLint,in vivo was calculated from in vivo pharmacokinetic data using three frequent mathematical models (the well stirred, parallel-tube, and dispersion models). The human scaling factor values (CLint,in vivo/CLint,in vitro) showed marked difference among the model compounds (0.3–26.6-fold). On the other hand, most of the animal scaling factors were within 2-fold of the values in humans, suggesting that scaling factor values were similar in the different animal species. When human CLint,in vitro values were compared with the actual CLint,in vivo, correlation was not necessarily good. By contrast, using human CLint,in vitrocorrected with the rat and/or dog scaling factors yielded better predictions of CLint,in vivo that were mostly within 2-fold of the actual values. Furthermore, successful predictions of human CLoral and hepatic extraction ratio (EH) were obtained by use of the human CLint,in vitro corrected with animal scaling factors. The new variant method is a simple one, incorporating additional information from animal studies and providing a more reliable prediction of human hepatic clearance.
In recent years, the process of drug discovery and development has become an increasingly time-consuming and costly endeavor. Much of the time and cost are expended on generating data that support the efficacy and safety profiles of the drug. Safe and efficient drug candidates must therefore be selected before clinical trials.
On the other hand, there is growing awareness of the key roles that pharmacokinetics and drug metabolism play as determinants of in vivo action. In these situations, early pharmacokinetic investigation plays an increasingly important role in the optimization and selection of drug candidates. In particular, it is important to predict human hepatic metabolic clearance because many drugs are eliminated from the body by hepatic metabolism. For predicting hepatic clearance, theoretical aspects of in vitro/in vivo scaling, based on a physiological model and clearance concepts, have been developed (Rane et al., 1977; Lin et al., 1982; Roberts and Rowland, 1986; Wilkinson, 1987). Application of this method has been successful in predicting in vivo hepatic clearance in rats for many drugs metabolized by P4501 from in vitro metabolism data using rat liver microsomes and isolated hepatocytes (Sugiyama et al., 1988;Houston, 1994). Since human liver samples have become more readily available, it would also be very useful to predict in vivo from in vitro data in humans. However, there has been relatively limited application of this approach (Hoener, 1994; Iwatsubo et al., 1997b), and there have been many failed attempts at predicting human hepatic clearance. For example, Iwatsubo et al. (1997a) reported a comparison of CLint,in vitro and CLint,in vivo for 25 metabolic reactions in humans from literature data. According to the report, although CLint,in vitro generally exhibited a positive correlation with CLint,in vivo, more than a 3-fold difference was observed in CLint for about 50% of the 25 metabolic reactions. Houston and Carlile (1997) also compared CLint,in vitro obtained from in vitro experiments using rat liver microsomes with CLint,in vivo for 28 drugs metabolized by P450. Although the predictability of CLint,in vivo from in vitro data was good overall, the results of some of the drugs tended to be low estimates. To improve the predictions of human hepatic clearance, a few investigators have described new methods and approaches. For example,Lave et al. (1997) have proposed the allometric scaling techniques combined with in vitro data. Obach (1999) has reported that inclusion of microsome binding values in the prediction of clearance from in vitro data appears to be a more broadly applicable approach.
In the present study, we have examined in vitro and in vivo metabolic clearance of eight model compounds, FK1052, FK480, zolpidem, omeprazole, nicardipine, nilvadipine, diazepam, and diltiazem, and calculated the CLint,in vitro and CLint,in vivo using in vitro and in vivo metabolism data in rats, dogs, and humans. At the same time, the measurement method of CLint,in vitro, which is determined from a substrate disappearance rate at 1 μM in hepatic microsomes, was used as a simple and useful method. We have also compared the parameters and evaluated a quantitative prediction method of human hepatic clearance focusing on P450 in drug discovery.
Materials and Methods
Chemicals.
FK1052, FK480, and nilvadipine were synthesized by Fujisawa Pharmaceutical Co., Ltd. (Osaka, Japan). Zolpidem hemitartrate and omeprazole sodium were kindly provided by Fujisawa-Synthelabo Pharmaceuticals (Tokyo, Japan) and Astra Japan, Ltd. (Osaka, Japan), respectively. Diltiazem hydrochloride and nicardipine hydrochloride were purchased from Sigma Chemical Co. (St. Louis, MO). Diazepam was purchased from Wako Pure Chemical Industries, Ltd. (Osaka, Japan). NADP+, glucose-6-phosphate, and glucose-6-phosphate dehydrogenase were obtained from Sigma Chemical Co. The other regents and solvents used were of analytical and HPLC grade.
Selection of Model Compounds.
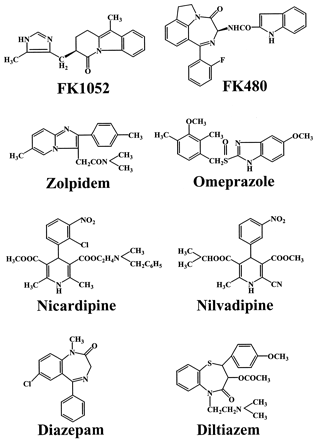
FK1052, FK480, omeprazole, zolpidem, nicardipine, nilvadipine, diltiazem, and diazepam (Fig. 1) were selected as the model compounds based on the following conditions:
-
Clearances of model compounds are determined by hepatic P450 metabolism
-
Extrahepatic clearances are assumed to be negligible
-
In vivo pharmacokinetic parameters in rats and/or dogs, and humans are reported
-
Absorption rates are good with no species difference
Chemical structures of model compounds.
Although it has been reported that diltiazem is metabolized in part by liver microsomal esterase in rats (LeBoeuf and Grech-Bélanger, 1987), the compound was examined for reference.
Hepatic Microsomes.
Liver specimens from adult male Sprague-Dawley rats (250–270 g,n = 3; Charles River Japan, Inc., Yokohama, Japan) and adult male dogs (9.5–10 kg, n = 3; Japan Laboratory Animals, Inc., Tokyo, Japan) were rinsed and homogenized with ice-cold 1.15% KCl. These pooled microsomes were prepared by differential centrifugation, and the 105,000g pellet was rinsed and resuspended in 1.15% KCl. Pooled human microsomes were obtained from Human Biologics International (Scottsdale, AZ). The pooled human microsomes were prepared from 15 individual liver donors that were selected on the basis of having average activities for the major P450 isozymes (1A2, 2A6, 2B6, 2C9, 2C19, 2D6, 2E1, 3A4, and 4A11). Each suspension was divided into aliquots, frozen, and stored at −80°C until used.
In Vitro Metabolism in Microsomes.
In vitro experiments
The time courses of the unchanged model compounds in microsomes were obtained. Each compound was incubated with a reaction mixture (500 μl) consisting of animal or human liver microsomal protein and NADPH-generating system (2 mM NADP+, 10 mM glucose-6-phosphate, 0.4 U/ml glucose-6-phosphate dehydrogenase, and 5 mM MgCl2) in the presence of 100 mM potassium phosphate buffer (pH 7.4). After preincubation at 37°C for 5 min, enzyme reactions were initiated by adding 5 μl of model compound solution in methanol. The final concentration of each model compound used was 1 μM. The microsomal concentrations used were 0.2 mg/ml (nicardipine, nilvadipine, and diltiazem), 0.5 mg/ml (FK1052, zolpidem, omeprazole, and diazepam), and 1.0 mg/ml (FK480). After incubation at 37°C for various time periods, the reactions of FK1052, FK480, omeprazole, zolpidem, nicardipine, and diltiazem were terminated by the addition of 500 μl of acetonitrile. The reactions of diazepam and nilvadipine were terminated by adding 500 μl of methanol and 3 ml of ethyl acetate, respectively. After stopping the metabolic reactions, the reaction mixtures of FK1052, FK480, omeprazole, zolpidem, nicardipine, diltiazem, and diazepam were centrifuged at 10,000g for 5 min, and an aliquot of the supernatant was injected on an HPLC for measuring the unchanged compound concentration. The reactions of nilvadipine were processed by extraction. The organic fraction was evaporated under N2, and the residue was reconstituted in the mobile phase (see below) for HPLC analysis.
Determination of unchanged model compound concentrations.
An LC module I plus (Millipore Co., Milford, MA) was used. The column for the analysis was an Inertsil ODS-3 (5 μm, 150 × 4.6 mm) (GL Science, Inc., Tokyo, Japan). The flow rate was 1.0 ml/min. The mobile phase and detection wavelength for the analysis of each model compound was as follows: FK1052, mobile phase: buffer A (5 mM phosphate buffer, pH 7.2)/CH3CN (50:50), detection: UV 242 nm; FK480, mobile phase: buffer A/CH3CN (40:60), detection: UV 295 nm; Zolpidem, mobile phase: buffer A/CH3CN (55:45), detection: UV 254 nm; Omeprazole, mobile phase: buffer A/CH3CN (60:40), detection: UV 302 nm; Nicardipine, mobile phase: buffer A/CH3CN (33:67), detection: UV 240 nm; Nilvadipine, mobile phase: buffer A/CH3CN (40:60), detection: UV 245 nm; Diazepam, mobile phase: buffer A/CH3OH (35:65), detection: UV 254 nm; Diltiazem, mobile phase: buffer A/CH3CN (50:50), detection: UV 240 nm.
All assay methods showed the concentration range of 0.1 to 2 μM. Reproducibility was evaluated by performing five replicate analyses of microsomal samples containing 0.1, 0.5, and 1 μM compound, respectively. The coefficient of variation was less than 10%, and the actual concentration of the compounds ranged from 88 to 112.1%. All assay methods thus provide good accuracy and precision.
Calculation of CLint,in vitro.
CLint,in vitro values were calculated from the substrate disappearance rate in hepatic microsomes as follows. If substrate disappearance can be assumed to follow a first-order reaction, the unchanged drug profile as a function of time [C(t)] is described as follows:
Furthermore, initial metabolic rate (V0) per unit milligram of microsomal protein (μmol/min/mg microsomal protein) is described by eq. 2.
On the other hand, from the Michaelis-Menten equation,V0 is described by eq. 3.
Physiological parameters for calculation of intrinsic clearance in rats, dogs, and humans
In Vivo Data.
Sources of pharmacokinetic data
In vivo clearance under linear conditions, fu and RB data, were obtained from in house and literature. The in vivo pharmacokinetic data were considered to be reliable since the in vivo pharmacokinetic experiments were performed based on accurate methods and appropriate protocols. The in vivo clearance value was calculated by dividing the dose by the area under the plasma concentration curve. When the in vivo clearance value was not expressed per kilogram of body weight, this value was converted so that it was expressed per kilogram of body weight by taking the mean value of body weight in the literature or a body weight of 250 g, 10 kg, and 70 kg for rats, dogs and humans, respectively. The fraction absorbed from the intestinal tract (Fa) of FK1052, FK480, and zolpidem were calculated by summing the recoveries of radioactivity in bile and urine after oral administration of14C model compounds to rats. Fa of omeprazole, nicardipine, nilvadipine, diazepam, and diltiazem were estimated to be 1.0 from the literature data.
Calculation of CLint,in vivo.
CLH values were determined from eq. 6 by use of the CLoral values, except the CLH for omeprazole in dogs, where only CLtot data was available. CLR was considered to be negligible for the model compounds.
EH were calculated from eq. 14.
Prediction of Human CLint,in vivo, CLoral, and EH.
Estimation of scaling factor
Values of scaling factor were estimated from the following equation:
Prediction of human CLint,in vivo.
Human CLint,in vivo were predicted based on human CLint,in vitro using the following two methods: 1) disregarding animal scaling factor:
Prediction of in vivo human CLoral and EH.
In vivo human CLoral and EHwere predicted using eqs. 6 to 14, based on the human CLint,in vitro corrected with animal scaling factor.
Binding of Model Compounds to Microsomes.
Determination of fu,microsome
Model compounds (final concentration, 1 μM) were mixed with liver microsomes (at protein concentrations used for the respective metabolic incubations) in 100 mM phosphate buffer (pH 7.4) containing 10 mM glucose-6-phosphate, 0.4 U/ml glucose-6-phosphate dehydrogenase, and 5 mM MgCl2. The mixtures (1.2 ml) were delivered to one side of a dialysis cell containing a preconditioned dialysis membrane (molecular weight cut off, 10 kDa) (Dainippon Pharmaceutical Co., Ltd., Osaka, Japan). To the other side of the membrane was delivered 1.2 ml of the phosphate buffer containing glucose-6-phosphate and MgCl2. The cells were sealed, and the apparatus was incubated at 37°C for 4 h. After an incubation period, the microsome and buffer samples were removed and analyzed by HPLC. The HPLC conditions were the same as described above. The unbound fraction was calculated from the following:
Calculation of CLuint,in vitro and the scaling factor.
By incorporating the correction with the unbound fraction in the microsomal incubation mixture, CLuint,in vitro is defined as (Iwatsubo et al., 1997a):
Results
In Vivo Pharmacokinetic Data of Model Compounds.
In vivo pharmacokinetic data for the model compounds are summarized in Table 2. Fa values were high, in the range of 0.7 to 1.0. The values of unbound fraction in plasma (or serum) (fp) were relatively low for all compounds ranging from the highest value for diltiazem (fp: rat, 0.184; dog, 0.298; human, 0.22) to the lowest value for FK480 (fp: rat, 0.01; dog, 0.006; human, 0.005). In vivo clearance, CLint,in vivo, and EH values differed markedly among the different species for each compound. Omeprazole, for example, is characterized by a high EH (0.94) in rats. An intermediate EH value (0.63) was observed in humans, whereas dogs exhibited a low EH (0.34). In vivo human EH ranged widely from 0.03 for diazepam to 0.87 for nilvadipine among the model compounds.
Estimation of CLint, in vivo from in vivo pharmacokinetic data for the model compounds in rats, dogs, and humans
In Vitro Metabolism in Microsomes and Estimation of Scaling Factor.
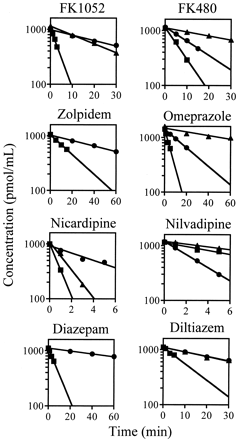
Figure 2 illustrates the time courses of the unchanged model compounds in microsomes. The unchanged drug profiles at 1 μM showed that the linear log concentration declines so that the metabolism follows first-order reaction under this condition. Large interspecies differences were also observed in CLint,in vitro for the model compounds. Table3 shows the human CLint,in vitro calculated from the time courses of the model compounds in microsomes and the values of human scaling factor, which are the ratios of CLint,in vivo obtained from in vivo pharmacokinetic data to CLint,in vitro. The human scaling factor values calculated using the well stirred model were about 26.6-fold for FK1052, about 5-fold for FK480, zolpidem, omeprazole, and nilvadipine, and 1- to 2.5-fold for nicardipine, diazepam, and diltiazem, showing marked difference among the model compounds. In the same way, the human scaling factor values calculated using the parallel-tube and the dispersion models were 0.3- to 16.1-fold and 0.4- to 18.2-fold, respectively, showing marked difference among the model compounds.
Time courses of unchanged compounds in microsomes.
Each compound (at a concentration 1 μM) was incubated for various time periods at 37°C in rat, dog, and human liver microsomes. Microsomal protein concentrations used were 0.2 mg/ml (nicardipine, nilvadipine, and diltiazem), 0.5 mg/ml (FK1052, zolpidem, omeprazole, and diazepam), and 1.0 mg/ml (FK480). ▪, rat liver microsomes; ▴, dog liver microsomes; ●, human liver microsomes. The solid lines represent the linear regression lines by the least-squares method.
Estimation of human CLint, in vitro in hepatic microsomes and their values of scaling factor for the model compounds
Figure 3 shows the differences in scaling factor between animals and humans. Most values of animal scaling factor were within 2-fold of the values in humans, except that the animal scaling factors for FK480 were 3.5- to 5.4-fold larger than that in humans. The results do not depend on the kind of the mathematical models and animal species.
Differences in scaling factor between animals and humans.
Symbols represent the fold differences of scaling factor calculated using the well stirred model (○), the parallel-tube model(▵), and the dispersion model(■). Closed symbols represent the fold differences of scaling factor between rats and humans. Open symbols represent the fold differences of scaling factor between dogs and humans. The solid line represents the line of unity. The area between the dotted lines represents an area within 2-fold error.
Prediction of Human CLint,in vivo, CLoraland EH by Use of Animal Scaling Factor.
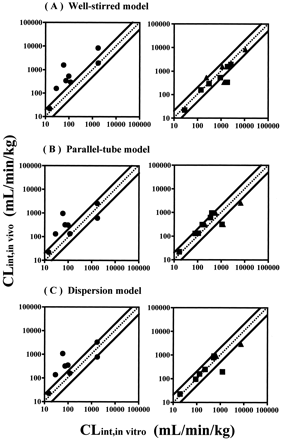
Human CLint,in vitro values corrected both with and without animal scaling factor are plotted versus actual CLint,in vivo values calculated using the mathematical models in Fig. 4. Without animal scaling factor consideration, only two (the well stirred model) or three (the parallel-tube and the dispersion models) of eight prediction values were within 2-fold of the actual CLint,in vivo, resulting in a large underestimation for most of the compounds. By contrast, using human CLint,in vitro corrected with rat and/or dog scaling factor, 9 (the parallel-tube model) or 10 (the well stirred and the dispersion models) of 13 prediction values were within 2-fold of the actual CLint,in vivo, significantly improving the predictability of CLint,in vivo.
Comparison of predicted CLint,in vitro with CLint,in vivo in humans calculated using the well stirred model (A), the parallel-tube model (B), and the dispersion model (C).
●, human CLint,in vitro not corrected by animal scaling factor using eq. 16; ▪, human CLint,in vitro corrected by rat scaling factor using eq. 17; ▴, human CLint,in vitrocorrected by dog scaling factor using eq. 17. The dotted lines represent the lines of unity. The area between the solid lines represents an area within 2-fold error.
The geometric mean accuracy values as indices of human CLint,in vivo predictability are listed in Table4. The value of one means perfect predictability, and the poorer becomes predictability with the larger values. The geometric mean accuracy values without animal scaling factor consideration were 3 to 4. By contrast, the predictability was substantially improved with the geometric mean accuracy value of less than 2.
Accuracy of human CLint, in vivo prediction methods
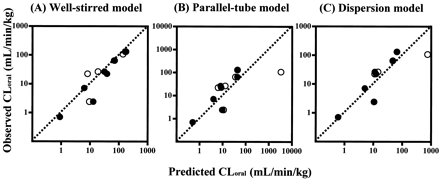
Furthermore, human in vivo CLoral and EH values were predicted from human CLint,in vitro corrected with animal scaling factor. Figure 5 presents comparisons of the predicted CLoral values with the observed CLoral. The CLoral values predicted using the well stirred model were in good agreement with the observed values. When CLoral was predicted using the parallel-tube and the dispersion models, good correlations were observed, with the exception of the overestimation for nilvadipine.
Comparison of predicted CLoralwith observed CLoral in humans calculated using the well stirred model (A), the parallel-tube model (B), and the dispersion model (C).
●, predicted CLoral calculated using human CLint,in vitro corrected by rat scaling factor; ○, predicted CLoral calculated using human CLint,in vitrocorrected by dog scaling factor. The dotted lines represent the lines of unity.
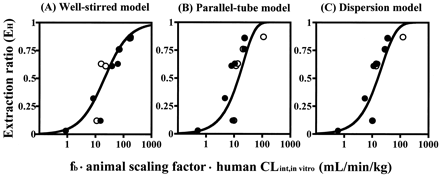
Figure 6 shows the relation between CLint,in vitro and in vivo EH. The predicted EH based on fb · CLint,in vitro corrected with animal scaling factor was close to the observed EH for all mathematical models. This result suggests that the human CLint,in vitrowith animal scaling factor consideration could give a good prediction of EH and hepatic clearance in humans.
Correlation between CLint,in vitro and in vivo EH in humans calculated using the well stirred model (A), the parallel-tube model (B), and the dispersion model (C).
●, human fb · CLint,in vitro corrected by rat scaling factor; ○, human fb · CLint,in vitro corrected by dog scaling factor. The solid lines represent the simulated curves based on the mathematical models.
Binding of Model Compounds to Microsomes.
Binding experiments were conducted in similar conditions used in in vitro microsomal metabolism studies but were conducted in the absence of NADP+ so that metabolism of the compounds would not occur. Recovery of the compounds was more than 90%, except nicardipine where the recovery was about 50%. Table5 summarizes the unbound fraction in liver microsomes, human CLuint,in vitro corrected with fu,microsome, and the scaling factor values. fu,microsome of diltiazem in rats could not be estimated because this compound was metabolized by microsomal esterase (see Materials and Methods). Human fu,microsome values were dependent on the model compounds. For example, although FK1052, FK480, nicardipine, and nilvadipine were highly bound to microsome with free fraction values ranging from 0.112 to 0.443, zolpidem, omeprazole, diazepam, and diltiazem showed low binding with free fraction values ranging from 0.745 to 0.975. fu,microsome of each compound to rat and dog liver microsomes was similar to that measured in human liver microsomes. For FK1052, FK480, and nilvadipine, the human scaling factor values obtained by correcting human CLint,in vitro with fu,microsome approached unity. However, the scaling factor for FK1052 were still 5.2- to 8.6-fold. In the case of nicardipine, these values by correcting human CLint,in vitro with fu,microsome became smaller than unity (0.05–0.1-fold). For zolpidem, omeprazole, diazepam, and diltiazem, corrections of CLint,in vitro with fu,microsome corrections did not change the human scaling factor values significantly. Consequently, the scaling factor for zolpidem and omeprazole remained 4.2- to 5.1-fold and 3.1- to 5.3-fold, respectively.
Binding of the model compounds to hepatic microsomes and their effects on predicting human CLint, in vivo
Discussion
In the present study, we investigated the quantitative prediction of human hepatic metabolic clearance from in vitro experiments using liver microsomes focusing on P450 metabolism. The values of CLint,in vitro and CLint,in vivo for each model compound were compared in rats and/or dogs, and humans. As a result, 1) scaling factor values (CLint,in vivo/CLint,in vitro) were similar in the different animal species; 2) scaling factor values were different in each compound; 3) successful predictions of human CLint,in vivo were obtained by considering animal scaling factor; and 4) use of human CLint,in vitro corrected with animal scaling factor gave good predictions of CLoral and EH in humans. Namely, the empirical prediction method is a simple one of incorporating additional information (which is compound specific) from animal studies and is based on the assumption that any in vitro-in vivo difference seen in humans is also apparent in animals to approximately the same degree.
In a conventional method, CLint,in vitro is obtained from Km andVmax, which are estimated by measuring the production of metabolites over a wide range of drug concentrations. On the other hand, CLint,in vitro in the proposed method is obtained from substrate disappearance rate at a single drug concentration. From comparison of these methods, advantages of the proposed method are raised as follows: 1) simple to conduct; 2) can be done for many compounds; 3) metabolites do not need to be known; 4) can be easily done without radiolabel; and 5) can yield enzyme kinetic data based on the disappearance of parent compounds. On the contrary, disadvantages of the proposed method are as follows: 1) it is difficult to measure very low CLint,in vitro values; 2) does not get individual metabolite information; 3) and does not obtainKm and Vmaxparameters. Recent studies have also calculated CLint,in vitro from substrate disappearance (Lave et al., 1997;Obach, 1999).
In this study, we assumed that extrahepatic clearances could be negligible. Recently, it has been reported that the first-pass metabolism in human small intestine is not negligible for some drugs, such as cyclosporine, which is metabolized mainly by CYP3A4 (Benet et al., 1996). Of the model compounds, FK480 (in house data), zolpidem (Pichard et al., 1995), nicardipine (Guengerich et al., 1991), nilvadipine (in house data), and diltiazem (Sutton et al., 1997) are metabolized mainly by CYP3A4. Human CLint,in vivoof these compounds were calculated from the CLoral after oral administration. Nevertheless, the CLint,in vivo values predicted from human CLint,in vitro with animal scaling factor consideration were comparable with the observed values (Fig. 4). From this result, although the possibility of intestinal metabolism of the model compounds cannot be completely excluded, it may be reasonable to consider the metabolism of these compounds mainly in the liver for predicting CLint,in vivo. In the future, we would evaluate in vitro-in vivo scaling for the model compounds obviously metabolized in small intestine.
When CLint,in vivo was calculated from in vivo clearance data, or when CLH and EH were calculated from CLint,in vitro, three frequent mathematical models (the well stirred, parallel-tube, and dispersion models) were used. Although there were a few drugs of which the human scaling factor values of all mathematical models were close to unity (nicardipine, diazepam, and diltiazem), the values for some drugs (FK1052, FK480, zolpidem, and omeprazole) were 3.1- to 26.6-fold (Table 3), resulting in CLint,in vivo values larger than the CLint,in vitro values. These findings suggest that the conventional prediction method, which directly applies CLint,in vitro into the mathematical models, cannot always predict in vivo clearance for all compounds. By contrast, the proposed method, considering the scaling factor, yielded more accurate prediction of human CLint,in vivo that was mostly within 2-fold of actual values (Fig. 4).
Presently, it is not clear why each compound has a intrinsic scaling factor. However, there are two possible reasons below. First, fu,microsome in the reaction mixture may have an influence on the CLint,in vitro value. Obach (1999) has examined fu,microsome of model compounds and the potential impact that such binding has on the prediction of in vivo clearance from CLint,in vitro for these compounds. Those results showed that incorporation of fu,microsome generally yielded more accurate predictions of human clearance. In the same way, we have evaluated fu,microsome of the model compounds and the change of the scaling factor values when correcting with fu,microsome. However, CLint,in vitro, when by correcting with fu,microsome, were not in agreement with the CLint,in vivo for some compounds; the scaling factor values for FK1052, zolpidem, and omeprazole were severalfold, and the scaling factor values for nicardipine were much smaller than unity (Table 5). This suggests that the scaling factor different from unity might not be due only to the drug binding to liver microsomes. The second reason may be related to some assumptions of the mathematical models below: 1) the distribution of drug into the liver is assumed to be perfusion-rate limited, and diffusion of drug into hepatocytes is rapid and not subject to any diffusional barriers; 2) it is assumed that only the free (unbound to macromolecules in blood) drug crosses the cell membrane and subsequently occupies the enzyme site; and 3) a homogeneous distribution of drug-metabolizing enzymes within the liver acinus is adopted (Houston and Carlile, 1997). If any assumptions are incorrect, the discrepancies between CLint,in vivo and CLint,in vitro would be observed.
The use of hepatic microsomes to predict in vivo CLH requires acceptance of some assumptions (Obach, 1999). First, oxidative metabolism predominates over other metabolic routes, such as direct conjugation, reduction, hydrolysis, etc. Second, rates of metabolism measured using animal and human microsomes in vitro are truly reflective of these exist in vivo. It has been reported that diltiazem is metabolized in part by hepatic microsomal esterase in rats (LeBoeuf and Bélanger, 1987). Therefore, the CLint,in vitro of microsomal esterase was estimated in conditions where no NADPH-generating system was included in the incubation mixture. As a result, the CLint,in vitro of microsomal esterase represented approximately 30% of the CLint,in vitro observed in the presence of NADPH-generating system in rats, whereas the CLint,in vitro of microsomal esterase in dogs and humans were hardly observed (data not shown). Even for diltiazem, scaling factor values were similar among the different species (Table 3), suggesting that the CLint,in vitro, which was measured in conditions with NADPH-generating in rats, reflects the metabolic rates by both P450 and esterase. However, future studies should also focus on evaluating in vitro-in vivo scaling for the other compounds metabolized by microsomal esterase to clarify the validity of the consideration. Third, the substrate concentration used (1 μM in this study) is well below the apparent Km. It may be necessary to confirm this assumption. For example, for only FK480, 3.5- to 5.4-fold differences in scaling factor between animals and humans were observed (Fig. 3). It may be accounted for by the possibility that the animal CLint,in vitro could be seriously underestimated because of the lower Kmvalue at 1 μM. However, the CLint,in vitro at several concentrations below 1 μM were similar to that at 1 μM in animals and humans (data not shown). Consequently, it was considered that the CLint,in vitro at 1 μM was under linear condition.
CLint,in vitro corrected with animal scaling factor could give a good prediction of in vivo CLoral and EH in humans. But, for a high-clearance drug, nilvadipine, the CLoral values using the parallel-tube and dispersion models were overestimated (Fig. 5). The reason is seen in the well stirred model where CLoral is in proportion to CLint,in vivo, whereas the parallel-tube and dispersion models give exponential correlation between CLint,in vivo and CLoral for high-clearance drugs (Fig.7). As a result, the error of predicted CLoral, which is caused from the error of predicted CLint,in vivo, is magnified in the parallel-tube and dispersion models compared with the error in the well stirred model. Also, in the oral case, CLint,in vivo calculated based on the parallel-tube and dispersion models was affected to some extent by QH for the high-clearance drugs, whereas CLint,in vivocalculated based on the well stirred model was not affected (Iwatsubo et al., 1997a). In the case of predicting CLoralfor high-clearance drugs, it may be necessary to consider the selection of the mathematical models.
Correlation between CLint and CLoral.
The solid line is the simulated curve using eqs. 6 to 8, based on the well stirred model. The dotted line is the simulated curve using eqs.6, 7, and 9, based on the parallel-tube model. The broken line is the simulated curve using eqs. 6, 7, and 10 to 13, based on the dispersion model.
In conclusion, we investigated a new variant method on the quantitative prediction of human hepatic clearance from in vitro experiments, focusing on P450 metabolism with eight model compounds. Successful predictions of human hepatic clearance were obtained by use of the human CLint,in vitro corrected with animal scaling factors, which are the ratios of CLint,in vivo to CLint,in vitro. This method would provide more reliable prediction of human hepatic clearance and be useful in drug discovery.
Footnotes
- Abbreviations used are::
- P450
- cytochrome P450
- HPLC
- high-performance liquid chromatography
- CL
- plasma clearance
- CLH
- hepatic clearance
- CLint
- intrinsic metabolic or hepatic clearance
- CLR
- renal clearance
- DN
- dispersion number
- EH
- hepatic extraction ratio
- Fa
- the fraction absorbed from the intestinal tract
- FH
- hepatic availability
- fp
- unbound fraction in plasma (or serum)
- fu,microsome
- unbound fraction in microsome
- QH
- hepatic blood flow rate
- RB
- blood-to-plasma concentration ratio
- Received February 20, 2001.
- Accepted July 3, 2001.
- The American Society for Pharmacology and Experimental Therapeutics