Abstract
Orally administered astemizole is well absorbed but undergoes an extensive first-pass metabolism toO-desmethylastemizole. Desmethylastemizole is formed in the human microsomal systems of the small intestine as well as the liver, which suggests the role of cytochromes P450 (P450s) in the first-pass metabolism of astemizole. Human P450s involved in theO-demethylation of astemizole have, however, not been identified, and the involvement of twelve known drug-metabolizing P450s were denied. During the course of the P450 identification study, higher activities of the astemizole O-demethylation in the rabbit small intestine than in the liver (about 3-fold) were found. These data suggest the possible involvement of CYP2J, since P450 included in this subfamily is dominantly expressed in the small intestine of rabbits. Therefore, CYP2J2 cDNA has been isolated from the human cDNA library and expressed in COS-1 cells. A clear activity of astemizole O-demethylation was detected in recombinant CYP2J2 with Km = 0.65 μM and Vmax = 1129 pmol/nmol P450/min. Expression of the immunoreactive protein with CYP2J2 antibody was detected in the small intestine and liver. Expression levels of the immunoreactive protein with the CYP2J2 antibody in the small intestine were well correlated with the activities of the astemizoleO-demethylation (r = 0.901,n = 5, p < 0.05). The CYP2J2 substrates, arachidonic acid and ebastine, strongly inhibited the microsomal astemizole O-demethylation in the human small intestines and recombinant CYP2J2. These results indicate the involvement of CYP2J2 in the presystemic elimination of astemizole in the human small intestine.
Oxidative metabolism of orally applied drugs during the first-pass is often critical for the pharmacological efficacy of these drugs. Several cytochrome P450 forms such as CYP3A4 eliminate portions of the drugs prior to reaching the circulation in the intestinal mucosa of humans (de Waziers et al., 1990; Kaminsky and Fasco, 1991; Kolars et al., 1992; 1994). Antihistamine drugs such as terfenadine (Garteiz et al., 1982), astemizole (Heykants et al., 1986), and ebastine (Yamaguchi et al., 1994) are extensively biotransformed in the small intestine and liver, and their first-pass oxidative biotransformations are essential for their pharmacological effect. In the case of terfenadine, the profound influence of grapefruit juice cointake indicates the major role of intestinal CYP3A4 on the first-pass metabolism (Benton et al., 1996; Honig et al., 1996; Rau et al., 1997). As we have recently reported, astemizole is mainly converted to the O-desmethyl derivative and other metabolites by multiple P450s1 in the human microsomal system (Fig.1; Matsumoto and Yamazoe, 2001). Similar to the case of terfenadine, the formation is likely to mainly occur in the small intestine. In contrast to the metabolism of terfenadine, no clear inhibition was, however, detected on the microsomalO-demethylation by the addition of the anti-CYP3A antibody and troleandomycin, a typical CYP3A4 inhibitor in the reaction mixture. Experiments using typical P450 form-specific inhibitors and antibodies did not give clear results for the participation of a specific P450 form in the human liver microsomal systems. In addition, none of the 12 commercially available recombinant P450s formedO-desmethylastemizole except for the negligible formation by the recombinant CYP2D6. Ebastine is also extensively biotransformed to its hydroxylated metabolite in the human small intestine, however, the P450 forms involving the ebastine biotransformation are still unidentified (Hashizume et al., 1998). A more recent report suggested that CYP4F12 and CYP2J2 may have catalytic activities toward ebastine (Hashizume et al., 2001; 2002), thus the possible involvement of these P450s may be speculated.
Proposed metabolic pathways of astemizole in humans
The CYP2J forms have been reported to be expressed in the small intestine of several experimental animals and humans (Scarborough et al., 1999). The CYP2J form was first isolated from the rabbit small intestine and was designated CYP2J1 (Ichihara et al., 1985; Kikuta et al., 1991). The human CYP2J member, CYP2J2, is shown to be expressed in several tissues such as the small intestine, liver, heart, kidney, and lung and may play a role in the oxidative bioactivation of arachidonic acid (Wu et al., 1996; Zeldin et al., 1996; 1997). However the properties such as the substrate specificity are not well characterized (Scarborough et al., 1999). In the present study, the microsomal metabolism of astemizole has been investigated using intestinal and liver microsomes from humans and rabbits. In addition, CYP2J2 cDNA has been isolated from the human liver and small intestinal cDNA libraries and expressed as the recombinant protein to evaluate the role of the astemizole metabolism. Based on these results, we have shown that CYP2J2 is involved in the intestinal first-pass metabolism of astemizole in humans.
Materials and Methods
Materials.
Individual human liver and intestinal microsomes were purchased from XenoTech LLC. (Cambridge, KS), BD Gentest Corp. (Woburn, MA) and Tissue Transformation Technologies (TTT, Edison, NJ). Rabbit liver and intestinal microsomes were prepared from male New Zealand white rabbits according to a previous report (Nakamura et al., 2000). Human intestinal and liver cDNA libraries were obtained from BD Biosciences Clontech (Palo Alto, CA). Arachidonic acid and phenytoin (Sigma-Aldrich, St. Louis, MO), astemizole and R55248 (Research Diagnostic Inc., Flanders, NJ) were purchased from the indicated sources. Ebastine and its hydroxylated metabolite were gifts from Dainippon Pharmaceutical Co., Ltd. (Osaka, Japan). The pCMV4 vector was provided by Dr. David W. Russels (University of Texas Southwestern Medical Center, Dallas, TX). The restriction endonucleases and DNA-labeling kit were purchased from Nippon Gene Co., Ltd. (Toyama, Japan). The nucleotide and [32P] were supplied from PerkinElmer Life Sciences (Boston, MA). The DNA ligation kit and Ex Taq were from Takara Shuzo Co., Ltd. (Kyoto, Japan). All other chemicals used were of the highest grade. The experiments with the human livers and intestines were approved by the Tohoku University Ethics Committee. The animal experiments were done under the instructions of the Tohoku University Animal Care and Use Committee.
Astemizole Metabolism by Human and Rabbit Microsomal Systems.
The incubation mixture for the astemizole metabolism consisted of 1 or 10 μM astemizole, 0.2 mg/ml microsomes, an NADPH-generating system (6.0 mM magnesium chloride, 3.3 mM glucose 6-phosphate, 1.3 mM NADP, 1.0 U/ml glucose 6-phosphate dehydrogenase) and 100 mM potassium phosphate buffer (pH 7.4) in a final volume of 0.2 ml. The reaction was initiated by the addition of the NADPH-generating system after 5 min of preincubation at 37°C and continued for 10 to 30 min. The reaction was terminated by the addition of 0.2 ml of acetonitrile. After the addition of R55248 as the internal standard (2 μM), the samples were centrifuged at 1800g for 5 min. Aliquots of the supernatant were analyzed by LC/MS. Astemizole was added as a methanolic solution in the reaction mixtures to ensure the final methanol concentration as less than 1%.
Kinetic studies were conducted in the presence of human small intestinal microsomes. All incubations were performed as previously described, except that the concentration of astemizole was varied (0.1, 0.2, 0.5, 0.75, 1, 2, 5, 10, 20, 50 μM). To avoid a secondary metabolism, the reaction was terminated after 10 min. In this condition, the metabolite concentration linearly increased depending on the incubation period. The kinetic data were fitted using the Michaelis-Menten model rate equation by nonlinear regression using WinNonlin (Scientific Consulting Inc., Version 1.5, Cary, NC) to estimate the Km andVmax values.
Cloning of CYP2J2.
PCR was carried out using a cDNA library from human liver as follows: Denaturation at 94°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 3 min, followed by 40 cycles. PCR primers used for amplifying the CYP2J-related sequences were designed based on the sequence of CYP2J2 cDNA (GenBank accession no. HSU37143). A λgt11 cDNA library of a human small intestine was screened with the32P-labeled PCR product. After the third screening, positive clones were isolated and purified. The DNA sequences of a clone were then separately determined using dye primers and Thermo Sequenase (Thermo Sequenase dye terminator cycle sequencing premix kit; Amersham Biosciences Inc., Piscataway, NJ) with an ABI373A DNA sequencer (Perkin-Elmer Japan, Tokyo, Japan). Since this first cDNA fragment lacked an initiation codon (seeResults), a pDR cDNA library of a human liver was further screened by the 32P-labeled cDNA fragment. After the third screening, a positive clone was isolated and purified. The DNA sequence of a clone was then separately determined using dye primers and Thermo Sequenase with DSQ-2000L (Shimadzu, Kyoto, Japan). Computer searches were performed using the BLAST program (http://www.ncbi.nlm.nih.gov/BLAST/). The designated CYP2J2 cDNA was ligated into a pCMV4 vector. The constructed plasmid DNA was transformed into COS-1 cells (Nagata et al., 1993). Harvested COS-1 cells with 0.25% trypsin containing 0.5 mM EDTA in phosphate-buffered saline were resuspended in HEPES-buffered saline (pH 7.5) at a cell density of about 5 × 106 cells/ml. Plasmid DNA was added to the suspended cells (80 μg of DNA/ml) and placed on ice for 10 min. Electroporation was carried out at 450 V and 250 μF using a Bio-Rad Gene Pulser (Bio-Rad, Hercules, CA) with the capacitance extender. After electroporation, the cells were immediately transferred to a 100-mm diameter Petri dish containing 10 ml of Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) with 10% fetal calf serum. The transformed COS-1 cells were cultured for 3 days in 10 ml of Dulbecco's modified Eagle's medium with 10% fetal calf serum under a humidified atmosphere of 95% air/5% CO2 at 37°C. The harvested COS-1 cells were sonicated and the microsomes were prepared by centrifugation for 1 h at 105,000g. The protein concentrations were measured by the method of Lowry et al. (1951) with bovine serum albumin as the standard. The microsomal contents of P450 were determined according to the method of Omura and Sato (1964). The designated CYP2J2 cDNA were also ligated into a prokaryotic expression vector, pQE30 (QIAGEN, Valencia, CA). The constructed plasmid DNA was transformed into the Escherichia coli, M15 strain. Recombinant protein was expressed and purified from bacteria by electrophoresis. Japanese white rabbits (2.2 kg, female) were intradermally immunized with 30 to 100 μg of purified CYP2J-E. coli protein in Hunter's Titer Max Adjuvant (Sigma-Aldrich). Two weeks after the boost, antisera were obtained and kept at −80°C until used.
Immunoblot Analysis.
Microsomal samples were subjected to immunoblot analyses using rabbit antisera raised against recombinant CYP2J-E. coli. Solubilized microsomes were separated by SDS-polyacrylamide gel electrophoresis with 8% acrylamide gels as previously reported (Laemmli, 1970). Each lane contains 50 μg of protein for the human intestinal and liver microsomes, 20 μg of protein for the rabbit intestinal and liver microsomes, and 20 μg of protein for the recombinant CYP2J2 microsomes. The separated proteins were electrophoretically transferred onto nitrocellulose filters (500 mA, 20 min). The protein was then visualized using peroxidase conjugated goat-antirabbit IgG (American Qualex, San Clemente, CA) and 3,3′-diaminobenzidine tetrahydrochloride as described elsewhere (Towbin et al., 1979). The developed band intensities were scanned by a flatbed scanner, and the data were processed by National Institutes of Health-image software (version 1.59). Correlation coefficients (r) between the band intensities and the activity of the astemizole O-demethylation were obtained after linear regression using Sigma Plot 2000 (SPSS Inc., Chicago, IL).
Astemizole O-Demethylation by Recombinant CYP2J2.
The incubation mixture for the astemizole O-demethylation or ebastine hydroxylation consisted of 100 μM of astemizole or ebastine, 0.2 mg/ml recombinant CYP2J2 microsomes, an NADPH-generating system (6.0 mM magnesium chloride, 10 mM glucose 6-phosphate, 1.0 mM NADP, 0.2 U/ml glucose 6-phosphate dehydrogenase) and 100 mM potassium phosphate buffer (pH 7.4) in a final volume of 0.2 to 0.5 ml. The reaction was initiated by the addition of the NADPH-generating system after 5 min of preincubation at 37°C and continued for 30 to 60 min. For astemizole, the samples were analyzed as described above. For ebastine, the reaction was terminated by the addition of 1 ml of ethyl acetate. Hydroxyebastine was extracted after the addition of phenytoin as the internal standard (100 μM), then evaporated to dryness. Aliquots of the solution (30 μl) were subjected to HPLC after the extract was redissolved in 100 μl of 20% acetonitrile.
Kinetic studies were conducted in the presence of the recombinant CYP2J2 microsomes. All incubations were performed as previously described, except that the concentration of astemizole (0.1, 0.2, 0.5, 0.75, 1, 1.5, 2, 5, 10 μM), and the amount of the recombinant CYP2J2 microsomes (0.2–2 mg/ml) were varied.
Inhibition.
For immunoinhibition, the anti-CYP2J2 antisera was preincubated with the human small intestinal microsomes or recombinant CYP2J2 microsomes at room temperature for 30 min. The incubation was started by the addition of astemizole (10 μM), the NADPH-generating system, and 100 mM potassium phosphate buffer (pH 7.4) in a final volume of 0.2 ml and continued for 30 min. The samples were processed and analyzed as previously described.
Arachidonic acid (10–200 μM) and ebastine (10–100 μM) were used as the chemical inhibitors. The incubation mixture consisted of astemizole (10 μM), an inhibitor, 0.2 mg/ml human small intestinal microsomes, the NADPH-generating system, and 100 mM potassium phosphate buffer (pH 7.4) in a final volume of 0.2 ml. The reaction was initiated by the addition of the NADPH-generating system after 5 min of preincubation at 37°C and continued for 20 to 30 min. The samples were processed and analyzed as described above.
LC/MS/MS and HPLC Apparatus.
For the LC/MS/MS analysis of the astemizole O-demethylation, a Hewlett-Packard 1050 HPLC system (Hewlett-Packard, Palo Alto, CA) and Finnigan MAT TSQ70 or TSQ700 were used (Thermo Finnigan, San Jose, CA). Separation of the desmethylastemizole was accomplished at 40°C in a reversed-phase mode using Develosil ODS-HG-5 (2.0 × 150 mm; Nomura Chemical Co., Ltd., Tokyo). The mobile phase consisted of acetonitrile/20 mM ammonium acetate in the proportions of 70:30. The flow rate was set at 0.2 ml/min. The metabolite and internal standard were detected in the positive ion electrospray ionization mode at the ion spray interface temperature of 200°C and identified using the transitions m/z 445→204 (O-desmethylastemizole), 475→475 (6-hydroxyastemizole), 325→325 (norastemizole) and 458→307 (R55248). The offset voltage was −30 V, and nitrogen and argon were used for the sheath and collision gases, respectively. The rate of the astemizoleO-demethylation was determined according to a concurrently generated standard curve.
For the HPLC analysis of the ebastine hydroxylation, a Jasco PU-980 pump system accompanying a JASCO DG-980-50 3-line degasser, JASCO LG-980-02 ternary gradient unit, Waters 712 WISP autosampler, JASCO UV-970 UV-VIS detector (254 nm) (Jasco, Tokyo, Japan) and SIC Chromatocorder 21J integrator (System Instruments, Tokyo, Japan) were used. Samples were separated at room temperature in the reversed-phase mode using a Capcellpak C18 (4.6 × 250 mm; Shiseido Co., Ltd., Tokyo, Japan). The mobile phase consisted of multiple gradients of 1% acetic acid and acetonitrile. After the injection of the sample, a linear gradient was started from 1% acetic acid/acetonitrile = 80:20 to 50:50 over a period of 30 min and continued for 8 min at the flow rate of 1.0 ml/min. The rate of ebastine hydroxylation was determined according to a concurrently generated standard curve.
Results
Astemizole O-Demethylation in Rabbit and Human Intestines and Livers.
The oxidative O-demethylation of astemizole was examined in systems containing microsomes from human livers, human small intestines, rabbit livers, and rabbit small intestines in the presence of 10 μM astemizole. Astemizole was mainly converted to the desmethyl derivative at the rate of 171 ± 57 pmol/mg of protein/min in the human small intestines and 478 ± 88 pmol/mg of protein/min in the human livers (mean ± S.D. of three samples). In the human small intestines, astemizole was also converted to norastemizole and 6-hydroxyastemizole at the rate of 30 ± 13 pmol/mg of protein/min and 13 ± 7 pmol/mg of protein/min, respectively. The apparent kinetic parameters of the astemizole O-demethylation using the human small intestinal microsomes wereKm = 1.8 ± 1.9 μM andVmax = 152 ± 86 pmol/mg of protein/min (Fig. 2; Table1). In the rabbit small intestines, astemizole was also mainly converted to the desmethyl derivative at the rate of 536 ± 257 pmol/mg of protein/min, which was roughly 2.9 times higher than the rate (185 ± 51 pmol/mg of protein/min) in the livers prepared from the same animal (mean ± S.D. of three samples).
S-V plots for astemizole O-demethylation in human small intestinal microsomes.
Astemizole (0.1, 0.2, 0.5, 0.75, 1, 2, 5, 10, 20, 50 μM) was incubated for 10 min with human intestinal microsomes (0.2 mg/ml). A, HJM0038; B, HJM0054; C, HJM0058; D, HJM0061; E, HJM0065. ●, observed data; solid lines, fitted curves.
Kinetic parameters for astemizole O-demethylation by human small intestinal microsomes
Expression of CYP2J2 in COS-1 Cells and Contents of CYP2J Protein in the Intestinal and Liver Microsomes.
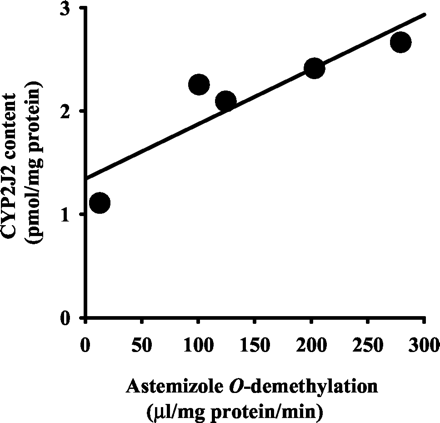
The nucleotide sequence of a clone obtained from the human small intestine and liver cDNA library was determined by dye terminator cycle sequencing. A GenBank database search using the BLAST program revealed that isolated cDNAs were matched to the sequence of CYP2J2 (GenBank accession no. NM000775.2). The cDNA obtained from the human small intestine was 1866 base pairs long (11–1519, open reading frame; 502, deduced amino acids) and no initiation codon was found in this clone. Therefore, these substitutions might be artificial modifications occurring during the construction of the cDNA library. The second clone obtained from a human liver cDNA library was 1850 base pairs long (6–1514, open reading frame; 502, deduced amino acids) and the nucleotide sequence had substitution in one nucleotide (C1021 to T) against the sequence of CYP2J2 (GenBank accession no. NM000775.2), although no difference was found in the deduced amino acid. This nucleotide sequence was deposited with the DDBJ nucleotide sequence database (accession no. AB080265). The liver cDNA was expressed in COS-1 cells using the mammalian expression vector pCMV4. In Western blotting with the antibody raised against CYP2J-E. coli, an immunoreactive protein band was detected at around 57 kDa (Fig.3). Typical CO-difference spectra with an absorption maximum at 450 nm were observed. The contents of the recombinant CYP2J2 in the microsomes of the COS-1 cells were 0.0053 nmol P450/mg of protein. Immunoreactive protein bands with the antibody were also detected at around 57 kDa in microsomes from the human small intestines and livers (Fig. 3A) and rabbit small intestines and livers (Fig. 3B). The immunoreactive band in the microsomal fractions prepared from the human small intestine was electrophoretically comigrated with the band in the recombinant CYP2J2 microsomes. The amount of CYP2J2 in the human small intestines was 2.1 ± 0.6 pmol/mg of protein (mean ± S.D., n = 5) using the recombinant CYP2J2 as the standard. The expression levels of CYP2J2 were well correlated with the astemizole O-demethylation (Fig.4; Y = 0.0053 X + 1.3435,r = 0.901, n = 5, p < 0.05). In human livers, the antibody against CYP2J2 immunoreacted with at least two electrophoretically distinct bands. A predominant band was electrophoretically similar to the immunoreactive band observed in the human small intestinal and recombinant CYP2J2 microsomes. Since this predominant band was not fully separated from another lower mobility band, the content of CYP2J2 in the livers could not be exactly evaluated. In rabbit, the specific amounts per mg protein were about 4.2 ± 1.1 times (Fig. 3B; mean ± S.D., n = 3) higher from the small intestine than from the liver.
Immunoblot analysis by anti-CYP2J2 antisera.
A, recombinant 2J2 microsomes (20 μg/lane); human small intestinal microsomes (50 μg/lane) (1, HJM0038; 2, HJM0054; 3, HJM0058; 4, HJM0061; 5, HJM0065; from TTT), human liver microsomes (6, pool of 16; 7, pool of 15; 8, H089; 9, H042; 10, H112, 6, 7 from Xenotech LLC.; 8, 9, 10 from BD Gentest Corp.). B, recombinant 2J2 microsomes, liver and small intestinal microsomes of three individual rabbits (20 μg/lane); control COS-1 microsomes (20 μg/lane). SI, small intestine; L, liver.
Simple regression analysis of the rate of astemizole O-demethylation and the expression levels of immunoreactive CYP2J2 protein in human intestines.
The rate of astemizole O-demethylation were indicated asVmax/Km values (μl/mg of protein/min; see Table 1). Lot HJM0038, HJM0054, HJM0058, HJM0061, and HJM0065 were used. ●, observed data; solid lines, fitted curves. Y = 0.0053 X + 1.3435, r = 0.901,n = 5, p < 0.05.
Activities of Recombinant CYP2J2.
The ebastine hydroxylation was determined to evaluate the catalytic properties of the recombinant CYP2J2 prepared from CYP2J2-expressing COS-1 cells in the presence of 100 μM ebastine. A significant catalytic activity was observed toward ebastine (5.5 nmol/nmol P450/min).
A clear activity was also observed for the astemizoleO-demethylation (0.4 nmol/nmol P450/min) with recombinant CYP2J2 in the presence of 100 μM astemizole. The activity decreased to 32 and 33% of the control with the addition of 50 μM ebastine and 100 μM arachidonic acid, respectively. The apparent kinetic parameters of the astemizole O-demethylation using recombinant CYP2J2 microsomes were Km= 0.65 μM and Vmax = 1129 pmol/nmol P450/min (mean of duplicate assays).
To assess the role of CYP2J2 on the metabolism of astemizole in the human small intestine, anti-CYP2J2 antisera was added to the systems containing the human small intestinal microsomes. Microsomal astemizoleO-demethylation was, however, not reduced by the addition of the anti-CYP2J2 antisera at 100 to 400 μl/ml. The astemizoleO-demethylation by recombinant CYP2J2 was also not diminished by the addition of the anti-CYP2J2 antisera at 200 μl/ml. Therefore, the antibodies raised against the purified CYP2J2 expressed in E. coli used were selective but judged not to be inhibitory.
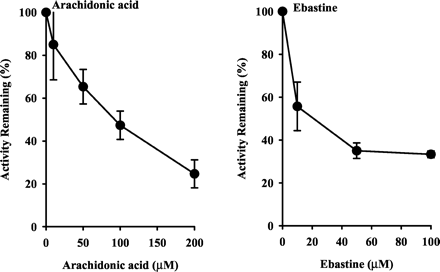
Inhibition by Arachidonic Acid and Ebastine.
Microsomal astemizole O-demethylation in small intestine from humans was examined in the presence of arachidonic acid and ebastine (Fig. 5). Both arachidonic acid and ebastine diminished the rate of microsomal astemizoleO-demethylation in concentration-dependent manners.
Effects of CYP2J2 substrates on astemizole O-demethylation by human small intestines.
Small intestine samples HJM0058, HJM0061, HJM0065 were used. Data are expressed as the percent activity remaining relative to a control (methanol alone) incubation and represented as mean ± S.D. of three samples.
Discussion
A clear catalytic activity for the astemizoleO-demethylation was detected in microsomes prepared from COS-1 cells expressing CYP2J2 cDNA. The expression of the immunoreactive protein in the human small intestines was detected and the expression levels were well correlated with the rate of astemizoleO-demethylation. The rate of astemizoleO-demethylation was clearly inhibited by the CYP2J2 substrates, arachidonic acid and ebastine (Wu et al., 1996; Zeldin et al., 1996; 1997; Hashizume et al., 2002). As we previously reported, astemizole O-demethylation was not detected using the commercially available twelve recombinant P450s except for the recombinant CYP2D6. However, none of the typical P450 form-specific inhibitors and antibodies including CYP2D6 inhibited the reaction in the human liver microsomal systems (Matsumoto and Yamazoe, 2001). Orally administered astemizole was well absorbed but underwent an extensive first-pass metabolism, thus the area under the curve of desmethylastemizole was about 20 times greater than that of astemizole (Heykants et al., 1986). The significant catalytic activity for the astemizole O-demethylation of the human small intestines suggest the role of P450s expressed in the small intestine in the first-pass metabolism of astemizole. During the course of the study, remarkable catalytic activities of the astemizoleO-demethylation were found in the rabbit small intestines. The astemizole O-demethylation was about 3 times higher than that in the livers. Several other marker activities of the P450 forms (phenytoin 4′-hydroxylation, dextromethorphanO-demethylation, testosterone 6β-hydroxylation) in rabbits were at least 4 times lower in the small intestine than in the livers (Nakamura et al., 2000). The CYP2J form was reported to be dominantly expressed in the rabbit small intestine, therefore, we focused on this P450 subfamily (Ichihara et al., 1985; Kikuta et al., 1991). The high expression of the CYP2J immunoreactive protein in the rabbit small intestine as compared to the liver was comparable to the catalytic properties of these tissues. The anti-CYP2J2 antisera used in this study had no inhibitory effect on the astemizoleO-demethylation in the human small intestines, in spite of the selective detection of the CYP2J protein in the Western blots. Although the reason remains obscure, the generated major epitopes are likely to be independent of the catalytic environment. The results from the chemical inhibition, expression level, and recombinant experiments strongly suggest that CYP2J2 is involved in the presystemic elimination of astemizole occurring in the human small intestine. In addition, significant expressed levels of CYP2J2 in the human small intestines and catalytic properties of CYP2J2 toward known substrates suggest the important role of CYP2J2 in the first-pass metabolism of therapeutic drugs.
The contribution of CYP2J2 to astemizole O-demethylation in the human liver has not yet been clarified. Although the expression of CYP2J2 in the human liver was detected by Western blotting, an additional band was detected with a slightly lower mobility. Since the immunoreactive band with a lower mobility partly overlapped the CYP2J2 band, the expression levels of CYP2J2 in the livers could not be determined. Involvement of another P450 in the astemizoleO-demethylation in the human liver cannot be negligible since a higher activity of astemizole O-demethylation was detected using human liver microsomes rather than human small intestinal microsomes. Another band with a lower mobility has not yet been determined. The antibody used in this study did not crossreact with other CYP2 family enzymes (2A6, 2B6, 2C9, 2C19, 2D6, 2E1) (unpublished data). Wu et al. (1996) also reported three electrophoretically distinct bands in microsomal fractions prepared from the human liver that immunoreact with the antibody raised against CYP2J2. These may represent alternative splice variants of CYP2J2, other uncharacterized human liver CYP2Js, or other P450s (Scarborough et al., 1999).
In the present study, a high expression of CYP2J in the rabbit small intestine and clear activity toward astemizoleO-demethylation in the rabbit small intestine were detected. Ebastine hydroxylation in the rabbit small intestines was also 1.5 times higher than that in the liver, which was comparable to the activity of the O-demethylation of astemizole (unpublished data). Since CYP2J1 was only found and dominantly expressing the CYP2J form in the rabbit small intestine, an immunoreactive protein band detected in the present study may correspond to CYP2J1 (Ichihara et al., 1985; Kikuta et al., 1991; Scarborough et al., 1999). The amino acid sequence of CYP2J1 has an 80% identity with that of CYP2J2, however, little is known about the catalytic activity of CYP2J1 except for the trace activity for benzphetamine, aminopyrine, andN,N-dimethylaniline (Scarborough et al., 1999). Our result may indicate that CYP2J1 catalyzes the oxidation of astemizole and ebastine in the rabbit small intestine. These results may also support the possibility that both astemizole and ebastine are the substrates of the CYP2J form in humans as well as in rabbits.
During the preparation of this manuscript, a paper was published on the CYP2J2 mediated metabolism of ebastine (Hashizume et al., 2002).
Acknowledgments
We thank Takahiro Nakamura (Biological Safety Research Center, National Institute of Health Sciences) and Kazuhiro Masubuchi (The Institute of Medical Science, The University of Tokyo) for their contribution to the cloning of CYP2J2.
Footnotes
- Abbreviations used are::
- P450
- cytochrome P450
- LC/MS
- liquid chromatography mass spectrometry
- MS/MS
- tandem mass spectometry
- PCR
- polymerase chain reaction
- HPLC
- high performance liquid chromatography
- Received April 23, 2002.
- Accepted August 8, 2002.
- The American Society for Pharmacology and Experimental Therapeutics