Interindividual Hepatic Expression, Substrate Specificity, and Role in Procarcinogen Activation
Abstract
The level of expression and interindividual variation in human hepatic microsomal cytochrome P450 (CYP) 2B6 was characterized using a polyclonal antibody (WB-2B6) raised against rat CYP2B1. Immunoblot analysis using cDNA-expressed human CYPs revealed strong cross-reactivity of this antibody with CYP2B6 (limit of detection < 0.05 pmol) and only minor cross-reactivities with human CYP2A6, CYP2D6, and CYP2E1, all of which could be resolved from CYP2B6 by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Analysis of human liver microsomes using this antibody revealed immunodetectable CYP2B6 protein in a majority of individual liver samples, with levels up to 74 pmol/mg protein in the CYP2B6-positive samples. Kinetic analysis of cDNA-expressed CYPs identified many of these enzymes as catalysts of 7-ethoxy-4-trifluoromethylcoumarin (7EFC)O-deethylation, but with significantly different apparentKM values (CYP1A2 < CYP2B6 ∼ CYP1A1 < CYP2C19 < CYP2C9 < CYP2E1 < CYP2A6). By assaying liver microsomal 7EFC O-deethylase activity at a low 7EFC concentration (5 μM) and preincubating human liver microsomes with anti-CYP1A, anti-CYP2C, and anti-CYP2E1 antibodies, we were able to monitor CYP2B6-dependent 7EFC O-deethylase activity in a panel of 17 human liver microsomes and observe a significant correlation (r2 = 0.80) between this activity and CYP2B6 protein content. The ability of CYP2B6 to activate prodrugs and procarcinogens was examined using gene locus mutation assays in CYP2B6-expressing human lymphoblast cells. CYP2B6-expressing cells were found to be more sensitive than control cells to the cytotoxicity and mutagenicity of cyclophosphamide, aflatoxin B1, and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. CYP2B6 is thus a widely expressed human liver microsomal CYP that can contribute to a broad range of drug metabolism and procarcinogen activation reactions.
CYPs1comprise a family of hemoproteins that are the principal enzymes involved in the biotransformation of many drugs and other foreign compounds, including anticancer drugs (1) and various promutagens and procarcinogens (2, 3). These enzymes are expressed in many mammalian tissues, with the highest levels found in liver. In recent years, considerable effort has been devoted to the study of human P450s, and this has resulted in the isolation, purification, and characterization of a number of these proteins. CYP3A,2CYP2C, and CYP1A2 are the most abundant P450 enzymes in human liver, comprising of ∼29%, 18%, and 13% of the total hepatic P450 content, respectively (4). Other forms of P450 found in human liver include CYP2E1 (5-7), CYP2A6 (8-10), CYP2D6 (11), and CYP2B6 (4, 12, 13).
Relatively little is known regarding the role of human hepatic microsomal CYP2B6 in xenobiotic metabolism. Studies with cDNA-expressed CYP2B6 have identified several substrates for this P450, including 7-ethoxycoumarin (12, 14, 15), benzo[a]pyrene (16), phenanthrene (17), 6-aminochrysene (13), methoxychlor (18), 1,3-butadiene (19), methoxyflurane (20), and 2-chloro-1,1-difluoroethene (21). CYP2B6 can also contribute to anticancer drug activation, as demonstrated by our recent observation that this P450 is a high KM catalyst of human hepatic microsomal cyclophosphamide 4-hydroxylation (15). Therefore, in high-dose cyclophosphamide chemotherapy administered in conjunction with autologous bone marrow transplantation (22), CYP2B6 may be an important enzyme in the activation of this alkylating anticancer prodrug. Substantial interindividual differences have been found in the contribution of CYP2B6 to cyclophosphamide 4-hydroxylation in a panel of human liver microsomes (15), and this may be related to the large intersubject variability in hepatic levels of CYP2B6 mRNA (12) and protein (4, 13). These differences are, in turn, consistent with the large interpatient differences reported in the clinical pharmacokinetics and biotransformation of cyclophosphamide (23-25). Currently, the underlying basis for the interindividual differences in hepatic CYP2B6 levels is not known. Detailed investigations of hepatic microsomal CYP2B6 have been limited, however, for several important reasons: a diagnostic catalytic marker for this P450 has not yet been identified; anti-CYP2B6 antibodies useful for Western blot analysis are not available; many of the heterologous anti-CYP2B antibody preparations are highly cross-reactive with other human P450 proteins; and a CYP2B6-specific chemical inhibitor has yet to be found.
The present study was undertaken to determine the level of CYP2B6 protein expression in a panel of human liver microsome samples using a CYP2B6-selective anti-rat CYP2B1 antibody (26), to evaluate microsomal 7EFC O-deethylase activity for its selectivity in monitoring hepatic CYP2B6, and to assess the role of this enzyme in the activation of several drugs and foreign chemicals. Our results show that, in striking contrast to previous reports (13, 17), CYP2B6 protein is present at a readily detectable level in a high proportion of human liver samples, as demonstrated using both immunochemical and enzymatic methods. Finally, CYP2B6-expressing human lymphoblasts were found to be sensitive to the induction of gene locus mutation by aflatoxin B1, NNK, and cyclophosphamide.
Materials and Methods
Reagents.
WB-2B6 is a whole serum preparation from a rabbit immunized with purified rat liver CYP2B1 (26). Where indicated, purified IgG fractions were prepared and used for immunoinhibition studies (e.g.see fig. 3). Anti-CYP2A6 and anti-CYP2E1 monoclonal antibodies, MAB-2A6 and MAB-2E1, respectively, were obtained from GENTEST Corp. (Woburn, MA). Anti-CYP1A2 antiserum (developed in rabbits, 69 mg/ml protein) and anti-CYP2C13 antiserum (developed in goats, 95 mg/ml protein) were purchased from Daiichi Pure Chemicals Co. (Tokyo, Japan). Microsomes containing individual human P450s expressed in human lymphoblasts were produced at GENTEST Corp. Nitrocellulose (0.45 μM pore) was purchased from Schleicher & Schuell, Inc. (Keene, NH). 7EFC and its 7-hydroxy metabolite 7-hydroxy-4-trifluoromethylcoumarin (7HFC), were purchased from Molecular Probes, Inc. (Eugene, OR) and Enzyme Systems Products (Livermore, CA). (S)-Mephenytoin was purchased from Ultrafine Chemicals (Manchester, UK). All other reagents were purchased from Sigma Chemical Co. (St. Louis, MO).
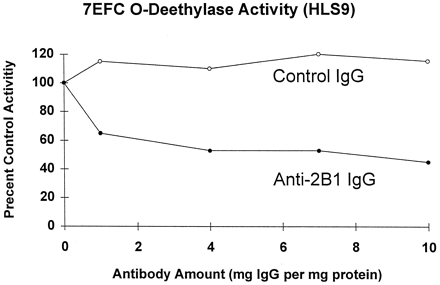
Inhibition of human liver microsome catalyzed 7EFC O-deethylase activity by anti-rat CYP2B1 IgG.
HLS9-catalyzed 7EFC metabolism was assayed for 30 min at 37°C using 25 μg microsomal protein and 50 μM 7EFC substrate in a total reaction volume of 0.5 ml. Included in each assay was the indicated amount of anti-CYP2B1 IgG or control rabbit IgG, expressed as a weight ratio to liver microsomal protein. Shown on the y-axis are activities relative to control incubations in the absence of antibody (100% = 437 pmol 7-hydroxy-4-trifluoromethylcoumarin formed/min/mg microsomal protein).
Human Liver Microsomes.
Human liver specimens obtained from organ donors after clinical death were kindly provided by Dr. A. Radominska (University of Arkansas for Medical Sciences, Little Rock, AR). Microsomes were prepared from individual liver samples, designated HLS2–HLS22, using methods described previously (27). Microsomal protein concentration was determined by the Bradford method with bovine serum albumin as standard.
CYP2B6 Immunoblot Analysis.
Samples were prepared by the addition of an equal volume of 2× SDS sample buffer [60 mM Tris (pH 6.8), 2% SDS, 20% glycerol, 2% of 2-mercaptoethanol, and 0.001% bromophenol blue], followed by heating for 5 min at 100°C. Samples were loaded on an SDS-PAGE and electrophoresed on a 14 × 16 cm 10% polyacrylamide gel at 60 V for 16–18 hr in a Hoefer electrophoresis apparatus. Proteins were transferred to nitrocellulose (0.45 μm pore size) by electroblotting at room temperature for 1 hr at 160 mA in 192 mM glycine, 25 mM Tris, and 20% methanol using a Hoefer Semiphor semidry transfer unit. The nitrocellulose was incubated in 4% powdered nonfat milk, 0.1% Tween 20, 25 mM Tris (pH 7.5), and 150 mM NaCl (blocking buffer) for 1 hr at room temperature. The nitrocellulose was incubated with WB-2B6 diluted 1:500 in blocking buffer for 1 hr at room temperature. The nitrocellulose was rinsed 3 times with blocking buffer for 5 min each wash. The nitrocellulose was incubated with alkaline phosphatase-conjugated rabbit anti-mouse IgG at 1:500 in blocking buffer, for 1 hr at room temperature. The nitrocellulose was rinsed 3 times with blocking buffer, then 2 times with 0.1% Tween 20, 25 mM Tris (pH 7.5), and 150 mM NaCl for 5 min each wash. The nitrocellulose was developed with nitroblue tetrazolium and 5-bromo-3-chloror-3-indolylphosphate developing solution at room temperature until bands appeared (5–15 min). The level of CYP2B6 protein in an individual human liver microsome sample was quantitated by blotting intensity. The blots were scanned using a flat bed scanner (Hewlett-Packard, Palo Alto, CA) and the bands quantified by use of Un-Scan It software (Silk Scientific, Inc., Orem, UT).
Enzyme Assays and Immunoinhibition Experiments.
7EFC O-deethylase activity was determined for each sample with and without preincubation (30 min on ice) with antibodies. Final antibody concentrations for anti-CYP2A6, CYP1A2, CYP2C13, and CYP2E1 were (in mg antibody/mg microsomal protein for the monoclonal antibody and ml antiserum/mg microsomal protein for the polyclonal antibodies) 0.75 mg/mg, 0.067 ml/mg, 0.3 ml/mg, and 0.33 mg/mg, respectively. Reaction mixtures containing 5 μM of 7-7EFC (unless indicated otherwise), 0.05 mg/ml microsomal protein, 1.3 mM NADP+, 3.3 mM glucose-6-phosphate, 0.4 units/ml glucose-6-phosphate dehydrogenase, and 3.3 mM magnesium chloride in 100 mM potassium phosphate (pH 7.4) were incubated at 37°C for 20 or 30 min in a total volume of 0.2 ml. Incubation was terminated by the addition of 40 μl of 20% trichloroacetic acid and then centrifuged (10,000 g) for 1 min. The supernatant (150 μl) was added to 1.85 ml of 100 mM Tris (pH 9), and fluorescence was determined by excitation at 410 nm and emission at 510 nm in a spectrofluorometer. Background fluorescence was subtracted from each unknown sample. 7EFC O-deethylase activity was quantitated by comparison with authentic 7-hydroxy-4-trifluoromethylcoumarin metabolite. Coumarin 7-hydroxylase activity was measured spectrofluorometrically (28). The final protein concentration was 0.05 mg/ml, and incubation time was 30 min. Diclofenac 4′-hydroxylase activity was measured according to Leemanet al. (29) at a final protein concentration of 0.2 mg/ml and an incubation time of 60 min. (S)-Mephenytoin 4′-hydroxylase activity was measured according to Wrighton et al. (30) at a final protein concentration of 1 mg/ml and an incubation time of 60 min. Phenacetin O-deethylase activity was measured according to Butler et al. (31) at a final protein concentration of 0.05 mg/ml and an incubation time of 30 min.p-Nitrophenol hydroxylase was measured according to Reinke and Moyer (32) at a final protein concentration of 0.25 mg/ml and an incubation time of 60 min. Taxol 6α-hydroxylase activity was measured according to Rahman et al. (33) at a final protein concentration of 0.25 mg/ml and an incubation time of 30 min. 7-Ethoxyresorufin O-deethylase activity was measured according to Burke et al. (34) at a final protein concentration of 0.05 mg/ml and an incubation time of 30 min. Testosterone 6β-hydroxylase was measured according to Waxman et al. (35) at a final protein concentration of 0.05 mg/ml and an incubation time of 30 min. Bufuralol 1′-hydroxylase was measured according to Kronbach et al. (36) at a final protein concentration of 0.05 mg/ml and an incubation time of 30 min.
Gene Locus Mutation Assays.
Mutation assays were conducted using a clonal, CYP2B6-expressing derivative of AHH-1 TK+/− human lymphoblasts (“h2B6 cells”) (15). h2B6 cells express CYP2B6 cDNA under control of the Herpes simplex virus thymidine kinase promoter in a vector conferring resistance to l-histidinol. Cells were maintained in RPMI medium 1640 supplemented to 9% with donor horse serum, containing 2 mMl-histidinol and without l-histidine. Gene locus mutation assays were conducted at the hprt locus as described (37) using h2B6 cells. These cells express CYP2B6 cDNA at a level of 50–60 pmol P450/mg microsomal protein. Cell concentration and treatment time were identical for all mutagenicity assays.
Results
Specificity of WB-2B6 Antibodies.
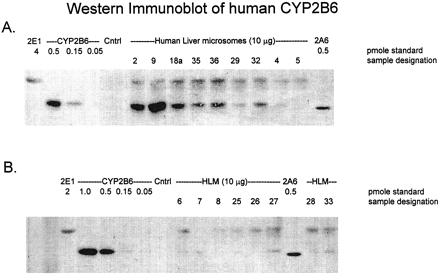
The specificity of WB-2B6 anti-CYP2B antibodies for individual human P450s was evaluated by immunoblot analysis using a panel of cDNA-expressed human CYPs. WB-2B6 gave a strong signal with 1 pmol CYP2B6 (fig. 1). Minor cross-reactivities were seen with CYP2A6, CYP2D6, and CYP2E1, which were detected at 15%, 2%, and 6% the sensitivity of CYP2B6 detection, respectively. Immunoreactive CYP2D6 and CYP2E1 proteins were well resolved from CYP2B6 based on their differences in SDS-PAGE mobility. The mobility difference between CYP2A6 and CYP2B6 was less, but complete resolution of these two CYPs could be achieved by use of a 10% acrylamide, 14 × 16 cm separating gel (fig. 2). WB-2B6 antibodies did not detect cDNA-expressed CYP1A1, CYP1A2, CYP2C8, CYP2C9, or CYP3A4 at 1 pmol (fig. 1) or 5 pmol P450 and CYP1B1, CYP2C19, and CYP4A11 at 2 pmol P450 (data not shown).
Western immunoblot analysis of cDNA-expressed P450 enzymes detected using WB-2B6 antibodies.
Gel loading was based on spectrophotometrically determined P450 contents (1 pmol/lane). CYP2B6 was found to give a strong signal, whereas weak cross-reactivities were detected with CYP2A6, CYP2D6, and CYP2E1. All cross-reacting bands could be resolved based on mobility in SDS-PAGE. Blotting was conducted as described in Materials and Methods. CON, control.
Western immunoblot analysis of human liver microsomes and microsomes prepared from human lymphoblastoid cells.
Human liver microsomes and WB-2B6 reactive, cDNA-expressed P450s were analyzed. The CYP2A6 standard demonstrates the difference in mobility for CYP2A6 and CYP2B6 under the conditions of this SDS-PAGE analysis. Identities and amounts of the samples loaded are indicated on the figure. Cntrl, control; HLM, human liver microsome.
Immunodetection of CYP2B6 in Human Liver Microsomes.
Immunoblot analysis of a panel of human liver microsomes revealed significant heterogeneity of CYP2B6 expression within the population of samples, with 12 of 17 individual samples found to contain ≥3 pmol CYP2B6/mg protein (fig. 2). The specific content of CYP2B6 ranged up to 74 pmol/mg microsomal protein in the CYP2B6-positive samples, as determined using spectrophotometrically quantitated, cDNA-expressed CYP2B6 as a standard. Using WB-2B6 antibodies, human liver microsomal CYP2E1 was detected in 16 of 17 samples, but the same antibodies did not detect CYP2D6 [perhaps due to the low expression level of CYP2D6 in human liver (4)] or CYP2A6 in this tissue. The human liver microsome samples that were very low in CYP2B6 all contained CYP2A6 as detected by coumarin 7-hydroxylase activity [mean activity: 775 pmol/(mg min)] and immunoblot analysis (using MAB-2A6 as a blotting reagent). (Note that whereas CYP2A6 standard is detected more strongly than in fig. 1, there is a difference in electrophoretic mobility of CYP2A6 standard and the human liver microsomal band detected in fig.2.)
Kinetic Analysis of 7EFC O-Deethylation by Human Liver Microsomes and cDNA-Expressed CYPs.
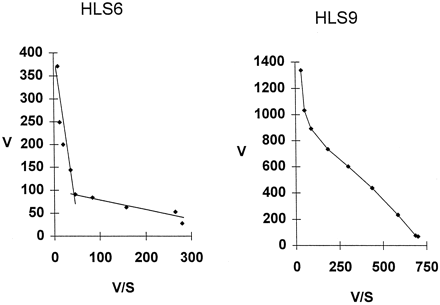
Eadie-Hofstee analysis of 7EFC O-deethylation catalyzed by a human liver microsome preparation is biphasic and suggests that at least two groups of enzymes contribute to this microsomal activity (38). One of these enzymes is likely to be CYP2B6, because as shown in fig. 3, anti-CYP2B IgG inhibited microsomal 7EFC activity partially, with a maximum of 60% inhibition found for HLS9-catalyzed 7EFC O-deethylation assayed at 50 μM substrate concentration. Indeed, comparison of the substrate concentration dependence of 7EFC activity in a CYP2B6-deficient liver sample, HLS6, to that of a CYP2B6-positive sample, HLS9, revealed both high KM (14 μM) and lowKM (<0.2 μM) components in HLS6 and a novel, intermediate KM component in HLS9 (fig.4).
Eadie-Hofstee plot of 7EFC O-deethylase activity for two human liver (HL) microsome samples: HLS6 and HLS9.
7EFC concentrations ranging from 0.1 to 40 μM were examined for liver samples HLS6 (deficient in CYP2B6, as determined by immunoblotting) and HLS9 (rich in CYP2B6, as determined by immunoblotting). V is expressed in pmol product/(mg protein × min). S is expressed in μM.
To identify the specific human P450s that are catalytically competent in 7EFC metabolism, and thus may potentially contribute to this activity in human liver, apparent KM values were determined for 7EFC O-deethylation catalyzed by each of the cDNA-expressed P450s. As summarized in table1, CYP1A2 is a very lowKM enzyme (apparent KM < 0.1 μM), CYP2E1 and CYP2C19 are high KM , high-capacity enzymes (apparent KM ’s of 46 μM and 17 μM, respectively), and CYP2A6 and CYP2C9 are highKM , low-capacity enzymes. By comparison, both CYP2B6 (apparent KM = 2.9 μM) and CYP1A1 (apparent KM = 1.8 μM) are intermediateKM catalysts of 7EFC O-deethylation. In contrast to the other enzymes listed in table 1, CYP1A1 is absent or expressed at very low levels in human liver (39-41) and thus is less likely to contribute to 7EFC metabolism in human liver.
Kinetic parameters for 7EFC O-deethylation: cDNA-expressed enzymes
CYP2B6 Component of Hepatic Microsomal 7EFCO-Deethylase Activity.
Application of the 7EFC O-deethylase assay (5 μM substrate) to the panel of 17 human liver samples revealed significant interindividual variation in activity levels with a mean activity of 222 pmol/(mg × min) (table 2). These activity measurements were correlated with immunoquantified CYP2B6 contents (r2 = 0.75; open symbols in fig. 5). However, the regression line did not pass close to the origin, indicating the presence of other enzymes contributing to this activity. Indeed, kinetic measurements with cDNA-expressed enzymes indicate that liver-expressed enzymes CYP1A2, CYP2C9/19, and CYP2E1 all have significant activity (table 1) and are likely to contribute to liver microsomal 7EFC O-deethylase activity at 5 μM substrate. To test this possibility, 7EFC O-deethylase activity was measured in the panel of 17 human liver microsomes both in the presence and absence of a mixture of anti-CYP1A, anti-CYP2C, and anti-CYP2E1 antibodies at saturating levels. Significant inhibition was observed with each liver microsomal sample (table 2). A residual mean activity of 108 pmol/(mg × min) was obtained. When CYP2B6-dependent 7EFCO-deethylase activity was determined by this approach, a better correlation (r2 = 0.80; closed symbols in fig. 5) was obtained between this enzyme activity and immunoquantified CYP2B6 content. In addition, the regression line now passed closer to the origin. Elimination of HLS2 and HLS9 from the data set (the two livers highest in CYP2B6) reduced the correlation coefficients (r2) for 5 μM 7EFC and 5 μM 7EFC plus antibodies to 0.23 and 0.62, respectively. The activity of cDNA-expressed CYP2B6 was essentially unaffected (84% activity) by the combination of three antibodies (table 2).
CYP2B6 component of 7EFC O-deethylase activity in a panel of human liver microsomes
Correlation of CYP2B6 immunoblotting intensity and the CYP2B6 component of 7EFC O-deethylase activity.
Immunoblotting/quantification and 7EFC O-deethylase activities were performed as described in Materials and Methods. Open symbols are data for 5 μM 7EFC (r2 = 0.75, top curve), and closed symbols are data for 5 μM 7EFC with the microsomes pretreated with anti-CYP1A2, anti-CYP2C13, and anti-CYP2E1 antibodies (r2 = 0.80, bottom curve).
Correlational Analysis of 7EFC O-Deethylase and P450 Enzyme-Selective Activities.
The observation that several livers that do not have detectable CYP2B6 protein (fig. 2) have a calculated CYP2B6 activity (table 2) suggests that this assay can detect other P450 form(s) that contribute to 7EFCO-deethylation in livers, whereas CYP2B6 is absent. To examine this possibility, we assayed the panel of human liver microsomal samples for catalytic activities associated with CYP1A (7-ethoxyresorufin O-deethylase and phenacetinO-deethylase) (40), CYP2A6 (coumarin 7-hydroxylase) (10), CYP2C8 (taxol 6α-hydroxylase) (33), CYP2C9 (diclofenac 4′-hydroxylase) (29), CYP2C19 [(S)-mephenytoin 4′-hydroxylase] (30), CYP2D6 (bufuralol 1′-hydroxylase) (42), CYP2E1 (p-nitrophenol hydroxylase) (43), and CYP3A (testosterone 6β-hydroxylase) (27, 44). Regression analysis indicated that the CYP2B6 component of 7EFC O-deethylase activity (determined as described previously) correlated (r2 = 0.78) with the CYP2A6-dependent coumarin 7-hydroxylase activity, but not with the other P450 enzyme-selective activities indicated previously.
Effect of Anti-CYP Antibodies on Human liver Microsomal 7EFCO-Deethylase Activity.
The regression analysis herein suggested that CYP2A6 may contribute to 7EFC O-deethylase activity in human liver. Therefore, immunoinhibition experiments were performed with anti-CYP antibodies to determine whether CYP2A6 contributes to 7EFC O-deethylase activity. An inhibitory MAB selective for CYP2A6 (designated MAB-2A6) did not significantly inhibit 7EFC O-deethylase activity in human liver microsome samples HLS2 and HLS9, which had high levels of both CYP2A6 and CYP2B6 protein. In control experiments, the same antibody inhibited coumarin 7-hydroxylase activity by >80% in these microsome samples (table 3). Thus, CYP2A6 does not significantly contribute to the CYP2B6 component of hepatic microsomal 7EFC O-deethylase activity. Anti-CYP1A, anti-CYP2C, and anti-CYP2E1 antibodies were partially inhibitory toward human liver microsomal 7EFC O-deethylase activity with the extent of inhibition varying with liver sample. In control experiments, these antibodies were 80–90% inhibitory their respective P450-enzyme selective catalytic activities (table 3).
Effect of antibodies on 7EFC O-deethylase and reference activities in human liver microsomes
Mutagenic Activation of Aflatoxin B1.
We have previously reported that aflatoxin B1 induces gene locus mutations in human B-lymphoblastoid cells that express human CYP1A1, CYP1A2, CYP2A6, or CYP3A4 (3, 28, 45). Because the present study showed that CYP2B6 is expressed in a high proportion of human liver samples, we examined the mutagenicity of aflatoxin B1in CYP2B6-expressing human B-lymphoblastoid cells (h2B6 cells). As shown in fig. 6, aflatoxin B1was mutagenic to h2B6 cells at exposure concentrations >50 ng/ml. In contrast, with control cells, aflatoxin B1 induces statistically significant mutagenicity only >1,000 ng/ml (45, 46). Therefore h2B6 cells are ∼20-fold more sensitive to aflatoxin B1 than control cells. Comparison of the relative efficiency (estimated by the product of minimum effective concentration and cellular P450 content; table 4) of CYP2B6 with other P450s in mutagenic activation of aflatoxin B1 indicated that the activity of CYP1A1, CYP1A2, CYP1B1, CYP2A6, and CYP3A4 with respect to activation of this promutagen are, respectively, 2.8, 140, 61, 5.4, and 14-fold higher than that of CYP2B6.
Mutagenicity of aflatoxin B1(closed circles), cyclophosphamide (CP; closed triangles), and NNK (closed squares) to h2B6 cells and mutagenicity of cyclophosphamide (open triangles) to control cells (untransfected AHH-1 TK+/− cells).
Mutagenicity data are plotted as the means of two independent experiments. Dotted line marks the 99% upper confidence limit (UCL) from the historical negative control data base at the hprt locus (a measure of statistical significance). The mean and SEM for the h2B6 cell negative controls were 1.7 × 10−6 and 0.4 × 10−6, respectively (N = 6). The mean and SE for the control cell negative controls were 1.6 × 10−6 and 0.4 × 10−6, respectively (N = 3).
Comparison of the abilities of cDNA-expressed P450 to activate aflatoxin B1 to a gene mutagen
Mutagenic Activation of NNK and Cyclophosphamide by CYP2B6.
Human CYP1A1, CYP1A2, CYP2A6, CYP2D6, and CYP2E1 are each capable of activating NNK (47-49), but whether CYP2B6 plays a significant role in this metabolic reaction is not known. In the present study, NNK was found to be mutagenic to h2B6 cells at exposure concentrations >5 μg/ml (fig. 6), whereas NNK was nonmutagenic to control cells at exposure concentrations up to 300 μg/ml (47). The relative efficiency of the different human P450 forms in NNK activation does not seem to vary as markedly as with aflatoxin B1. After correcting for P450 content (using the same previous approach as used for aflatoxin B1), the ability to activate metabolically NNK is approximately CYP2B6 = CYP1A2 = CYP2A6 > CYP2E1 = CYP2D6. Therefore, CYP2B6 seems to be one of several human P450s that are comparatively efficient with respect to NNK activation.
Cyclophosphamide is cytotoxic to CYP2B6-expressing human B-lymphoblastoid cells in culture (15). In the present study, we examined the mutagenicity of CYP2B6 in the same cell line. As shown in fig. 6, cyclophosphamide was found to be mutagenic at exposure concentrations >50 μg/ml, whereas CYP2B6-negative control cells were not sensitive to cyclophosphamide at exposure concentrations up to 150 μg/ml. This is consistent with a role of CYP2B6 in cyclophosphamide activation.
Discussion
Previous studies have reported large intersubject variability in hepatic levels of CYP2B6 mRNA (12) and protein (4, 13). The levels of CYP2B6 protein detected in the panel of human liver microsomes examined in the present study are, however, substantially higher than that those reported previously (4). CYP2B6 apoprotein was detected in the majority of human liver microsome samples examined. The levels of CYP2B6 expression were quite heterogeneous among different microsome samples and ranged up to 74 pmol CYP2B6/mg microsomal protein for the CYP2B6-positive samples. Conceivably, the low level of human liver microsomal CYP2B6 reported previously may be due to the use of a heterologous antibody with poor cross reactivity with human CYP2B6. Alternatively, it may reflect variability due to sample size or ethnic differences in hepatic CYP2B6 expression. According to Shimada et al (4), CYP2B6 was detected in only 30% of Japanese liver samples (N = 30), but could be detected in 85% of Caucasian liver samples (N = 30). Moreover, the average hepatic CYP2B6 protein content was ∼10-fold higher in the Caucasian samples (1.4 ± 1.8 pmol/mg microsomal protein; N = 32) than in the Japanese samples (0.14 ± 0.62 pmol/mg microsomal protein; N = 42) (50). Indeed, many of the liver samples included in the present study were obtained from Caucasian donors (table 2). Finally, the differences in apparent CYP2B6 contents between these two studies may result from differences in tissue processing. In the course of the present studies we noted that the CYP2B6 catalytic activity for one of most enriched samples (HLS9) decreased markedly upon storage on ice. Therefore, it is possible that CYP2B6 exhibits a higher lability to degradation than other P450 forms. Additional studies are needed to clarify this point.
In agreement with a previous report (38), a nonlinear Eadie-Hofstee plot was obtained for 7EFC O-deethylation by human liver microsomes, suggesting that multiple P450s can catalyze this reaction. Consistent with this proposal, cDNA-expressed CYP1A1, CYP1A2, CYP2A6, CYP2B6, CYP2C9, CYP2C19, and CYP2E1 were each shown to be catalytically active in metabolizing 7EFC, but with widely different apparentKM values (CYP1A2 < CYP1A1 ∼ CYP2B6 < CYP2C19 < CYP2C9 < CYP2E1 < CYP2A6). CYP1A2 exhibited a very low KM (<0.1 μM), suggesting this P450 is the high-affinity 7EFC O-deethylase in human liver. Both CYP1A1 and CYP2B6 have intermediate apparentKM values for 7EFC O-deethylation. However, it is likely that CYP2B6, rather than CYP1A1, represents the novel, intermediate KM form of 7EFCO-deethylase found in sample HLS9 because CYP1A1 is absent or expressed at very low levels in human liver (39-41). CYP2C19, CYP2C9, CYP2E1, and CYP2A6 have relatively high apparentKM values for 7EFC O-deethylation, suggesting that these four P450s correspond to the low-affinity 7EFCO-deethylases in human liver. However, based on immunoinhibition experiments and kinetic parameters as determined with cDNA-expressed enzymes, CYP1A2 and CYP2E1 are, in addition to CYP2B6, likely to be significant contributors to liver microsomal 7EFC activity at 5 μM substrate. CYP2C19 may also contribute to overall activity at 5 μM 7EFC in microsome samples rich in this enzyme. CYP2C9 is likely of limited importance given its slow rate of 7EFC metabolism, its high apparent KM , and the fact that the buffer conditions of the 7EFC assay (0.1 M potassium phosphate) markedly inhibit CYP2C9 (51). Using this information, we were able to develop an antibody mixture approach that can estimate the CYP2B6 component of human hepatic microsomal 7EFC O-deethylase activity. Based on this method, the levels of CYP2B6-associated microsomal 7EFCO-deethylase activity were shown to be correlated with hepatic CYP2B6 protein levels with a regression line passing close to the origin. There may be a small “residual 7EFCO-deethylase activity representing the 10–20% of the competing enzymes that are not inhibited by the antibody mixture.
Although coumarin 7-hydroxylase activity correlated with the CYP2B6 component of human liver microsomal 7EFC O-deethylase activity, CYP2A6 does not contribute to the CYP2B6 component of 7EFCO-deethylase activity, as indicated by the observation that a MAB (MAB-2A6) selective for CYP2A6 does not inhibit 7EFCO-deethylase activity in human liver microsomes. The correlation of CYP2B6 activity and CYP2A6 activity may be due to coordinate regulation of the two respective genes that has been reported by others (52).
Results from the present investigation in conjunction with those of previous studies (46, 49, 53) indicate that multiple P450s can activate aflatoxin B1 (CYP1A1, CYP1A2, CYP1B1, CYP2A6, CYP2B6, and CYP3A4). Aflatoxin B1 has a low mutagenic potency in CYP2B6-expressing cells relative to that in cells expressing CYP1A1 (49), CYP1A2 (53), CYP2A6 (46), and CYP3A4 (53). This is indicated by its minimum effective mutagenic concentration and by its relative efficiency for mutagen activation [calculated as the product of minimum effective concentration (A in table 4) and cellular P450 content (B in table 4)]. Given CYP2B6’s low efficiency of aflatoxin activation and its low specific content in human liver (this study and ref. 4), hepatic CYP2B6 is likely to play only a minor role in aflatoxin B1 activation. Indeed, immunoinhibition and chemical inhibition experiments have indicated that CYP1A2 and CYP3A form(s) are the major P450(s) responsible for activation of this promutagen in human liver (54-56).
The tobacco-specific nitrosamine NNK requires activation in vivo to produce mutagenicity, as demonstrated in a rodent model (57). Several individual cDNA-expressed human P450s have been identified as catalysts of NNK activation in vitro, including CYP1A1, CYP1A2, CYP1B1, CYP2A6, CYP2D6, and CYP2E1 (47-49). In the present study, NNK was found to be mutagenic to CYP2B6-expressing human B-lymphoblastoid cells in culture. In contrast to aflatoxin B1, NNK was efficiently activated to mutagenic species by CYP2B6, when compared with CYP1A2, CYP1B1, CYP2A6, CYP2E1, and CYP2D6 (47, 48). Although NNK can be metabolized to two DNA-adducting intermediates [a methylating agent and a pyridyloxobutylating agent (57, 58)], it remains to be established whether CYP2B6 is involved in one or both of these metabolic pathways. Recent studies have reported that NNK can be activated by both human liver and lung microsomes (48, 59). In human liver, CYP2A6 and CYP2E1 seem to be important P450 catalysts of NNK activation (59). Currently, it is not known whether human hepatic CYP2B6 contributes significantly to NNK activation.
Cyclophosphamide was presently shown to be mutagenic to h2B6 cells, and this is consistent with our previous finding that CYP2B6 is one of several human P450s that can activate this cancer chemotherapeutic agent by 4-hydroxylation (15). Among a panel of 11 purified rat liver P450 enzymes, CYP2B1 is the most active catalyst of this reaction (60) and, not surprisingly, cyclophosphamide is mutagenic to CYP2B1-expressing V79 Chinese hamster cells (61) and to CYP2B1-expressing Saccharomyces cerevisiae (62). The range of exposure concentration required for cyclophosphamide mutagenicity seems to be similar for these systems. Whether hepatic CYP2B6 is important in the mutagenicity of cyclophosphamide is not known, but hepatic CYP2B6 is a high KM catalyst of the 4-hydroxylation of this drug (15). In a recent study, we identified allelic variants of CYP2C9 and the polymorphically expressed CYP2C19, as lower KM enzymes for this drug activation reaction (63). Further studies are required to compare the relative efficiency of CYP2B6 and the CYP2C forms in mutagenic activation of cyclophosphamide.
In summary, CYP2B6 was expressed in a majority of the human liver samples examined and at levels that are substantially higher than those reported previously (4, 50). Multiple human hepatic P450 enzymes were found to be catalytically competent for 7EFC deethylation and enzyme kinetic analysis identified CYP2B6 as an intermediateKM enzyme for this reaction. A method using a low 7EFC concentration and a mixture of inhibitory antibodies was developed that provides a reasonable correlation between microsomal 7EFC O-deethylase activity and hepatic CYP2B6 levels. Finally, CYP2B6 was shown to be active in the mutagenic activation of aflatoxin B1, NNK, and cyclophosphamide in CYP2B6-expressing human B-lymphoblasts in culture.
Footnotes
-
Send reprint requests to: Dr. Charles L. Crespi, GENTEST Corporation, 6 Henshaw Street, Woburn, MA 01801.
-
This study was supported in part by the National Institutes of Health Grant CA-49248 (to D.J.W.) and, in part, by the National Institutes of Health Grant P42-ES-04675-10 (GENTEST).
-
↵2 Individual cytochrome P450 forms are designated according to the systematic nomenclature (3b).
- Abbreviations used are::
- CYP
- cytochrome P450
- P450
- cytochrome P450
- 7EFC
- 7-ethoxy-4-trifluoromethylcoumarin
- NNK
- 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone
- IgG
- immunoglobulin G
- MAB
- monoclonal antibody
- HLS
- human liver microsomal sample, with each individual liver sample identified by number (e.g. HLS6, HLS9, etc.)
- SDS
- sodium dodecyl sulfate
- SDS-PAGE
- sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- hrpt
- hypoxanthine phosphoribosyltransferase
- Received December 18, 1996.
- Accepted April 9, 1997.
- The American Society for Pharmacology and Experimental Therapeutics
References
- 1.↵
- 2.↵
- 3a.
- 3b.
- 4.↵
- 5.↵
- 6.↵
- 7.↵
- 8.↵
- 9.↵
- 10.↵
- 11.↵
- 12.↵
- 13.↵
- 14.↵
- 15.↵
- 16.↵
- 17.↵
- 18.↵
- 19.↵
- 20.↵
- 21.↵
- 22.↵
- 23.↵
- 24.↵
- 25.↵
- 26.↵
- 27.↵
- 28.↵
- 29.↵
- 30.↵
- 31.↵
- 32.↵
- 33.↵
- 34.↵
- 35.↵
- 36.↵
- 37.↵
- 38.↵
- 39.↵
- 40.↵
- 41.↵
- 42.↵
- 43.↵
- 44.↵
- 45.↵
- 46.↵
- 47.↵
- 48.↵
- 49.↵
- 50.↵
- 51.↵
- 52.↵
- 53.↵
- 54.↵
- 55.↵
- 56.↵
- 57.↵
- 58.↵
- 59.↵
- 60.↵
- 61.↵
- 62.↵
- 63.↵