Abstract
The contributions of specific human liver cytochrome P-450 (CYP) enzymes to the activation, via 4-hydroxylation, of the oxazaphosphorine anticancer prodrugs cyclophosphamide (CPA) and ifosfamide (IFA) were investigated. Analysis of a panel of 15 human P-450 cDNAs expressed in human lymphoblasts and/or baculovirus-infected insect cells (Supersomes) demonstrated that CYPs 2A6, 2B6, 3A4, 3A5, and three CYP2C enzymes (2C9, 2C18, 2C19) exhibited significant oxazaphosphorine 4-hydroxylase activity, with 2B6 and 3A4 displaying the highest activity toward CPA and IFA, respectively. CYP2B6 metabolized CPA at a ∼16-fold higher in vitro intrinsic clearance (apparentVmax/Km) than IFA, whereas 3A4 demonstrated ∼2-fold higherVmax/Km toward IFA. A relative substrate-activity factor (RSF)-based method was developed to calculate the contributions of individual P-450s to total human liver microsomal metabolism based on cDNA-expressed P-450 activity data and measurements of the liver microsomal activity of each P-450 form. Using this method, excellent correlations were obtained when comparing measured versus predicted (calculated) microsomal 4-hydroxylase activities for both CPA (r = 0.96,p < .001) and IFA (r = 0.90,p < .001) in a panel of 17 livers. The RSF method identified CYP2B6 as a major CPA 4-hydroxylase and CYP3A4 as the dominant IFA 4-hydroxylase in the majority of livers, with CYPs 2C9 and 2A6 making more minor contributions. These predicted P-450 enzyme contributions were verified using an inhibitory monoclonal antibody for 2B6 and the P-450 form-specific chemical inhibitors troleandomycin for 3A4 and sulfaphenazole for 2C9, thus validating the RSF approach. Finally, Western blot analysis using anti-2B6 monoclonal antibody demonstrated the presence of 2B6 protein at a readily detectable level in all but one of 17 livers. These data further establish the significance of human liver CYP2B6 for the activation of the clinically important cancer chemotherapeutic prodrug CPA.
Cyclophosphamide (CPA)1and ifosfamide (IFA) are isomeric cancer chemotherapeutic prodrugs that are bioactivated by specific cytochrome P-450 (CYP) enzymes (Clarke and Waxman, 1989) to form therapeutically active, cytotoxic metabolites (Sladek, 1988). P-450 metabolism of these drugs yields a primary 4-hydroxy metabolite that equilibrates with the ring-opened tautomer aldophosphamide, which, in turn, undergoes chemical decomposition to yield a bifunctional mustard derivative (phosphoramide or ifosphoramide mustard) and acrolein. The mustard metabolite generates a highly electrophilic aziridinium species that forms DNA cross-links, proposed to be the key cytotoxic lesions induced in tumors treated with oxazaphosphorines (Sladek, 1988; Fleming, 1997). Alternatively, the 4-hydroxy metabolite can be deactivated by aldehyde dehydrogenase to yield the therapeutically inactive carboxyphosphamide (Sladek, 1993). Intratumoral expression of aldehyde dehydrogenase activity is an important mechanism of resistance to this class of drugs (Sladek, 1993;Bunting and Townsend, 1996).
Studies with isolated liver microsomes have established that overlapping subsets of liver P-450 enzymes catalyze the activation of CPA and IFA. Both oxazaphosphorines are activated by constitutive P-450 enzymes belonging to the 2C subfamily and also by drug-inducible P-450s belonging to subfamilies 2B and 3A in both the rat model (Clarke and Waxman, 1989; Weber and Waxman, 1993) and in humans (Chang et al., 1993). 2B6 and 3A4 were proposed to correspond to the highKm catalysts of CPA and IFA 4-hydroxylation, respectively, detected in human liver microsomes (Chang et al., 1993) whereas a subset of the CYP subfamily 2C enzymes [e.g., 2C19 (Chang et al., 1997a)] exhibits lowKm values and is proposed to contribute to the low Km oxazaphosphorine 4-hydroxylase activity component seen in this tissue (Chang et al., 1993, 1997a).
Large interpatient differences in the clinical pharmacokinetics and biotransformation of CPA (Chen et al., 1995; Yule et al., 1996) and IFA have been reported (Boddy et al., 1996). Rodent model studies demonstrate that alterations in liver P-450 enzyme compositions and activities can have a major impact on the pharmacokinetics of CPA and IFA metabolism (Brain et al., 1998; Yu et al., 1999), suggesting that the interindividual differences in P-450-dependent oxazaphosphorine pharmacokinetics seen in the clinic may, in part, reflect individual differences in liver P-450 enzyme profiles. These pharmacokinetic differences may be of therapeutic significance (Ayash et al., 1992) and, consequently, it is important to develop a more complete understanding of the manner in which interindividual differences in human hepatic P-450 enzyme profiles (Shimada et al., 1994) influence the capacity of individual human livers for CPA and IFA 4-hydroxylation. Because cancer patients are very often treated with multiple drug regimens, the identification of specific P-450s involved in the bioactivation of CPA and IFA may enable clinicians to predict, and thereby avoid, potential drug-drug interactions that might compromise therapeutic efficacy. In addition, the identification of polymorphically expressed members of P-450 subfamily 2C (Goldstein and de Morais, 1994), which demonstrate significant differences in CPA, and IFA 4-hydroxylase activity (Chang et al., 1997a) could potentially contribute to some of the variations in patient response toward oxazaphosphorine treatment. This knowledge could, in turn, lead to the design of therapeutic strategies to maximize oxazaphosphorine drug activation and limit systemic toxicity or potential drug-drug interactions.
In the present study, we have analyzed CPA and IFA activation catalyzed by 15 individual human P-450s expressed in either human lymphoblastoid cell-derived microsomes or baculovirus-infected insect cell microsomes. By using P-450 form-selective antibody and chemical inhibitors, we have demonstrated an unambiguous role for CYPs 2B6 and 3A4 in human liver microsomal oxazaphosphorine activation. Importantly, a major role for P-450 2B6 in human liver CPA 4-hydroxylation was established, in contrast to a recent study that failed to demonstrate the participation of P-450 2B6 in human hepatic microsomal CPA activation (Ren et al., 1997). Finally, the measurement of human liver microsomal P-450 profiles has enabled us to apply a substrate-activity based method to predict overall rates of microsomal drug metabolism and the contributions of individual P-450 enzymes to the drug metabolic reaction under investigation. Using this method, the principal human liver microsomal P-450 CPA and IFA 4-hydroxylases were identified and their relative catalytic contributions toward overall hepatic metabolism of these anticancer prodrugs were calculated.
Materials and Methods
Chemicals.
CPA and IFA were obtained from the Drug Synthesis and Chemistry Branch, National Cancer Institute (Bethesda, MD). 4-Hydroperoxy-CPA was a gift from Dr. J. Pohl (ASTA Pharma, Bielefeld, Germany). Troleandomycin (TAO) was obtained from Pfizer Inc. (Brooklyn, NY). NADPH and sulfaphenazole were purchased from Sigma Chemical Co. (St. Louis. MO). All other specialty chemicals were obtained from Aldrich Chemical Co. (Milwaukee, WI). Lymphoblast microsomes and Supersomes containing cDNA-expressed human P-450s and monoclonal antibody specific to 2B6 (MAB-2B6; Yang et al., 1998) were obtained from Gentest Corporation (Woburn, MA).
cDNA-Expressed Human P-450s.
Microsomes containing cDNA-expressed human P-450s were from the AHH-1 TK−/+ human B lymphoblastoid cell line (“lymphoblast P-450s”) or baculovirus-infected insect cells (“Supersomes”) engineered to express individual human P-450 cDNAs as well as human P-450 reductase cDNA. Some of the Supersomes also contained human cytochrome b5, which was cDNA-expressed in the baculovirus system with the concentration of cytochrome b5determined spectrally. Specific catalytic activities exhibited by lymphoblast P-450s were, in general, comparable to those exhibited by typical human liver samples, whereas the Supersomes generally exhibited specific P-450 catalytic activities severalfold higher than the average human liver microsomal sample (HLS) when calculated on a per mg protein basis. Negative controls corresponding to lymphoblast or Supersome microsomes without coexpressed P-450s were run in parallel in each enzymatic incubation.
Human Liver Microsomes.
Human liver specimens obtained from organ donors after clinical death were kindly provided by Dr. A. Radominska (University of Arkansas for Medical Sciences, Little Rock, AR). Microsomes were prepared from individual liver samples, designated HLS2 to HLS36, using methods described previously (Waxman et al., 1988). Microsomal protein concentrations were determined by the Bradford method with bovine serum albumin as standard.
CPA and IFA 4-hydroxylase Assay.
The fluorometric determination of microsomal CPA and IFA 4-hydroxylase activity was performed as described (Weber and Waxman, 1993) with minor modifications. Incubation mixtures typically contained either 0.25 or 2.0 mM CPA or IFA, 1 mM NADPH, 5 mM semicarbazide hydrochloride and either 100 μg human liver microsomal protein or 20 to 40 pmol of cDNA-expressed P-450 protein in a total volume of 0.1 ml of 100 mM potassium phosphate buffer (pH 7.4) containing 0.1 mM EDTA. Complete incubation mixtures minus NADPH were preincubated for 4 min at 37°C. Reactions were initiated by the addition of 1 mM NADPH and incubated a further 60 min at 37°C. Reactions were terminated by the sequential addition of 40 μl of ice-cold 5.5% (w/v) zinc sulfate, 40 μl of ice-cold saturated barium hydroxide, and 20 μl of ice-cold 0.01 M hydrochloric acid. After centrifugation for 15 min at 16,000g, 0.15 ml of the supernatant was transferred to a clean test tube containing 80 μl of fluorescence reagent (6 mg of 3-aminophenol and 6 mg of hydroxylamine hydrochloride dissolved in 1 ml of 1 M hydrochloric acid). Samples were heated at 90°C for 20 min in the dark and then allowed to cool to room temperature before reading the fluorescence (excitation at 350 nm and emission at 515 nm) on a Shimadzu RF-1501 spectrofluorophotometer. Standard curves for acrolein were generated by incubating 4-hydroperoxy-CPA (0–20 μM) with bovine serum albumin in parallel under the same assay conditions.
Relative Substrate-Activity Factor (RSF) Method for Calculation of P-450 Enzyme Contributions to Human Liver Microsomal Metabolism.
A method was adapted from the relative activity factor (RAF) approach described previously (Crespi, 1995) to integrate data from cDNA-expressed CYPs to determine the relative contributions of individual P-450 enzymes to a given P-450 reaction (“test substrate” activity) in human liver microsomes. A panel of diagnostic P-450 substrates (P-450 form-specific substrates; see Table 2 legend) was first used to assay the catalytic activities of each of ten different human CYPs, expressed in both lymphoblast microsomes and Supersomes. The panel of human liver microsomes whose test substrate activities were to be predicted was also assayed with each of the diagnostic P-450 substrates. The cDNA-expressed P-450s were also assayed for the test substrate activities under investigation, in this case CPA and IFA 4-hydroxylation (2 mM substrate). An RSF was then calculated for each cDNA-expressed P-450 by dividing its catalytic activity with the test substrate by its catalytic activity with its diagnostic substrate (eq. 1). For example, for CYP3A4 Supersomes, RSFIFA (4-OH-IFA/6β-OH-testosterone) = 4.72/124 = 0.038. The RSF value of each lymphoblast or Supersome P-450 preparation indicates how much more active (RSF value >1) or how much less active (RSF value <1) the cDNA-expressed enzyme preparation is with the test substrate compared with the diagnostic P-450 substrate when each substrate is assayed under its own respective assay conditions. Because the same cDNA-expressed enzyme preparation is used to assay both substrates, results obtained using the RSF values are, to a first approximation, independent of the factors that influence the absolute enzyme activity of any given cDNA-expressed P-450 preparation, factors that often vary from one cDNA-expressed P-450 to another. These factors include the specific P-450 protein content, the level of endogenous or expressed P-450 reductase, and the level of cytochrome b5. The RSF values thus obtained were used to calculate the test substrate activities (e.g., CPA and IFA 4-hydroxylase activities) attributed to each P-450 form in a given HLS by multiplying the measured liver microsomal rate of metabolism of each diagnostic substrate (representing a specific P-450 form) by the RSF value corresponding to that same P-450 form (eq. 2). For example, for HLS2, the testosterone 6β-hydroxylation rate of 4890 pmol/min/mg × RSFIFA value of 0.038 gives a predicted 3A4-dependent IFA 4-hydroxylase activity in HLS2 of 186 pmol/min/mg (see example shown in Table 2). The sum of RSF-derived microsomal rates of metabolism for all ten P-450 forms gives the total calculated (i.e., predicted) rate of metabolism of the test substrate for that liver microsome sample (eq. 3). The RSF-derived rate of liver microsomal metabolism by each specific P-450 form divided by the total calculated activity of the liver microsome gives the calculated contribution of that P-450 form to total hepatic microsomal metabolism (eq. 4). For example, 186/456 = 40.8% of total predicted HLS2 microsomal IFA 4-hydroxylase activity calculated as being associated with P-450 3A4.
RSF-derived CPA and IFA 4-hydroxylase activities of different P-450 forms in HLS2
The RSF-based approach described above can be applied to the test substrate reaction CPA (or IFA) 4-hydroxylation using the following equations, where P-450x represents a specific P-450 form and liver n represents a single human liver microsomal preparation.
Predicted total 4-hydroxylation rate by liver microsomen = Sum of calculated microsomal 4-hydroxylation rates by all liver P-450s in liver microsome n (eq. 3)
Chemical and Antibody Inhibition Studies.
TAO, freshly dissolved in methanol, was added to individual assay tubes and the solvent was then evaporated using a SpeedVac concentrator. The TAO residue was dissolved in 4-hydroxylase assay buffer containing the complete assay mixture minus substrate (CPA or IFA). The samples were then preincubated for 30 min to allow for the formation of CYP3A inhibitory TAO metabolites. The 4-hydroxylase assay was subsequently initiated by the addition of CPA or IFA, followed by a fresh aliquot of 1 mM NADPH in a final assay volume of 0.1 ml. Control incubations included methanol vehicle alone, to control for the effects of the 30-min preincubation with NADPH, which decreased 4-hydroxylation activity by up to ∼30 to 40% compared to assays in the absence of TAO (Yu and Waxman, 1996). Experiments with other chemical inhibitors were performed without the preincubation protocol. Immunoinhibition experiments with MAB-2B6 (10 mg/ml stock; 0–10 μl dissolved in a total of 10 μl of 25 mM Tris buffer, pH 7.4) included a 30-min preincubation step of the antibody with microsomes at room temperature. Control incubations were performed with microsomes and Tris buffer without antibody.
CYP2B6 Immunoblot Analysis of Human Liver Microsomes.
Microsomes (40 μg protein) were resolved on 7.5% SDS gels, which were then electrotransferred to nitrocellulose blots using standard methods. Blots were rinsed in Solution I (10 mM PBS, pH 7.4 containing 0.05% Tween 20) for 5 min, then blocked with 3% powdered nonfat dry milk dissolved in Solution II (10 mM PBS, pH 7.4 containing 0.3% Tween 20) for 1 h at room temperature. The blots were then washed (5 min/wash) in Solution I. MAB-2B6 diluted 1:1000 (v/v) in Solution I containing 3% powdered nonfat dry milk was then incubated with the nitrocellulose blot for 1 h at room temperature while shaking, followed by two 5-min washes of the blot in Solution I. The nitrocellulose was then probed with secondary antibody [horseradish peroxidase-conjugated sheep anti-mouse IgG diluted 1:3000 (v/v) in Solution I] for 1 h at room temperature. The blot was washed with Solution II followed by Solution I (15 min/wash) and developed with ECL reagent (Amersham) until clear bands appeared on X-ray film (∼3 min). Blots were scanned by a densitometer and the level of 2B6 protein was quantified by comparison to lymphoblast-expressed 2B6 protein standard (0.13–1.0 pmol/lane).
Results
Role of Individual cDNA-Expressed Human CYPs in CPA and IFA Activation.
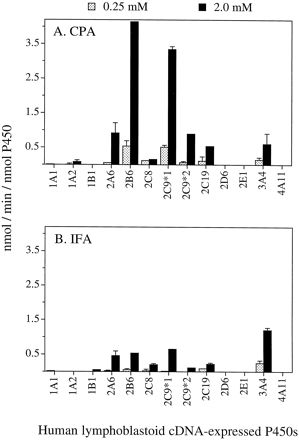
Two different P-450 expression systems were used to identify human P-450 enzymes that are catalytically competent in the 4-hydroxylation of CPA and IFA: microsomes prepared from human B lymphoblastoid cells (“lymphoblasts”) and microsomes prepared from baculovirus-infected insect cells (Supersomes). Human P-450 reductase cDNA was coexpressed with each P-450 cDNA in both expression systems. In the lymphoblast system, CYPs 2A6, 2B6, 2C9, 2C19, and 3A4 showed significant oxazaphosphorine 4-hydroxylase activity, whereas CYPs 1A1, 1A2, 1B1, 2C8, 2D6, 2E1, and 4A11 exhibited little or no activity (Fig.1). Similar activity patterns were obtained at low (0.25 mM) and high (2.0 mM) substrate concentrations. However, several of the 4-hydroxylation activities were too low for reliable quantitation at 0.25 mM substrate; hence, we have restricted our data analysis and discussion to results obtained at 2 mM substrate except as noted. With CPA as substrate, CYPs 2B6 and 2C9-Arg144 allele (2C9*1, the most abundant 2C9 allele) showed the highest catalytic activity, whereas with IFA as substrate, CYP3A4 was the most active, followed by 2A6, 2B6, and 2C9*1 (Fig. 1B). All of the active P-450s except 3A4 showed higher 4-hydroxylase activity with CPA compared with IFA. P-450s 2B6 and 2C9*1 were ∼5- to 7-fold more active with CPA compared with IFA, whereas 3A4 was ∼2-fold more active with IFA compared with CPA.
Activation of CPA (A) and IFA (B) at 0.25 and 2.0 mM drug concentration catalyzed by human lymphoblast cDNA-expressed CYPs.
Each of the cDNA-expressed P-450s contains coexpressed human NADPH P-450 reductase. Data are expressed as mean ± S.E.M. (n = 3 determinations).
Cytochrome b5 is expressed endogenously in lymphoblasts but not in the insect cell host for the Supersome system. Because cytochrome b5 can play a facilitatory role in several P-450-catalyzed reactions, including 3A4 (Yamazaki et al., 1996), the effect of cytochrome b5 was investigated in the Supersome expression system (Table1). In the case of 3A4, inclusion of cytochrome b5 at a 1.5:1 molar ratio to P-450 had no effect on CPA or IFA 4-hydroxylase activity (Table 1A, line 3 versus line 2). By contrast, significant increases in 4-hydroxylase activity were obtained with increasing P-450 reductase levels, both with and without cytochrome b5 (Table 1A, line 1 versus 2 and line 3 versus 4). For each of the other P-450s, the available Supersome preparations that contained cytochrome b5 also contained higher levels of P-450 reductase (Table 1), making it difficult to determine with certainty whether the observed increase in enzyme activity was due to the presence of cytochrome b5 or to the increase in P-450 reductase activity. However, the increases in the 4-hydroxylase activities of Supersomes 2C8, 2C9*1, 2C19, and 3A5 in the presence of cytochrome b5 each appear to be somewhat larger than would be expected based on the fold increase in P-450 reductase alone (e.g., ∼5-fold increase in 2C9 4-hydroxylase activity with only a 2.5-fold increase in the Supersome’s P-450 reductase content; Table1B), suggesting that cytochrome b5 does stimulate the 4-hydroxylase activities of these latter P-450s. In contrast, the 4-hydroxylase activity of 3A7 was not stimulated by cytochrome b5 (Table 1B).
Effect of cytochrome b5 and NADPH P-450 reductase on CPA and IFA 4-hydroxylase activities of Supersomes
Comparison of the CPA and IFA 4-hydroxylase activities of the full panel of Supersomes, comprised of 15 different human P-450s, revealed that 2B6 exhibited the highest CPA 4-hydroxylase activity followed by 2C19, 3A4, 3A5, 2C9*1, 2A6, and 2C18. Other P-450s (1A1, 1A2, 1B1, 2C8, two alleles of 2C9 [2C9-Cys144 allele (2C9*2) and 2C9-Leu359 allele (2C9*3)], 2D6, 2E1, 3A7, and 4A11) all showed lower, or in many cases no, detectable CPA 4-hydroxylase activity. In contrast, IFA 4-hydroxylase activity was highest with 3A4 followed by 2C19 and 3A5 (Fig. 2). Of the active P-450s available in the Supersome panel but unavailable in the lymphoblast panel, 2C18 showed ∼3-fold higher 4-hydroxylase activity with CPA compared with IFA, whereas 3A5 was ∼1.5-fold more active with CPA. Comparison of the two P-450 cDNA expression systems revealed that 2B6 and 3A4 were both ∼3- to 4-fold more active per nmol P-450 in insect cells than in the lymphoblast expression system. In spite of these differences, however, both cDNA expression systems demonstrated 2B6 and 3A4 to be the most active P-450 enzymes toward CPA and IFA activation, respectively.
4-Hydroxylation of CPA (A) and IFA (B) at 0.25 and 2.0 mM drug concentration catalyzed by human P-450 cDNAs expressed in Supersomes.
Each of the cDNA-expressed P-450s contains coexpressed human NADPH P-450 reductase. P-450s 2A6, 2B6, 2C8, 2C9*1, 2C19, 2E1, 3A4, 3A5, and 3A7 also contain cytochrome b5 at a cytochrome b5/P-450 M ratio of 2.5, 2.2, 1.6, 1.8, 1.5, 1.0, 1.8, 1.5, and 0.9, respectively (also see Table 1). Data are expressed as mean ± S.E.M. (n = 3 determinations).
Kinetic Analysis of Oxazaphosphorine 4-Hydroxylation by cDNA-Expressed CYPs.
2B6 and 3A4 exhibit severalfold higher 4-hydroxylase activities at 2 mM compared with 0.25 mM substrate for both CPA and IFA (Figs. 1 and 2), suggesting that they may correspond to highKm oxazaphosphorine 4-hydroxylases. Apparent Km andVmax values for these reactions were therefore determined by steady-state enzyme kinetics (Fig.3). 2B6 demonstratedKm values of 2 to 3 mM toward CPA and IFA, whereas 3A4 showed a ∼2-fold lower Kmwith IFA (∼0.8 mM) as compared to CPA (∼1.8 mM; Fig. 3A).Vmax values for 3A4 ranged from 7 to 8 nmol/min/nmol P-450 toward both substrates. In contrast, 2B6 exhibited ∼10-fold higher apparent Vmax toward CPA compared with IFA (Fig. 3B). As a result, the catalytic efficiency of 2B6, judged by in vitro intrinsic clearance (apparentVmax/Km), was ∼16-fold higher for CPA than IFA. By comparison, 3A4 exhibited an intrinsic clearance for IFA that was ∼2-fold higher than for CPA (Fig. 3C). TheseVmax/Km ratios indicate that 2B6 exhibits a strong kinetic preference for CPA activation, whereas 3A4 exhibits a more modest preference for IFA activation.
Enzyme kinetic analysis of CPA and IFA 4-hydroxylation catalyzed by Supersomes 2B6 and 3A4.
The apparent Km (A) andVmax (B) values were based on 4-hydroxylation activities for Supersomes containing cytochrome b5 assayed at substrate concentrations ranging from 0.25 to 5 mM and were calculated using the software program Enzyme Kinetics (ChemSW, Inc., Fairfield, CA). In vitro intrinsic clearance values (C) was calculated by dividing averageVmax by averageKm values. Data are shown as mean ± ½ range of duplicate determinations.
Human Liver Microsomal Oxazaphosphorine 4-Hydroxylation.
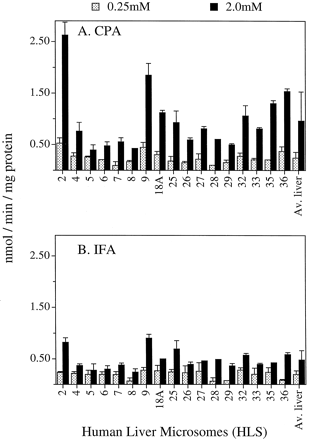
A panel of 17 human liver microsomes was assayed for 4-hydroxylation of CPA and IFA (Fig. 4). Assays were carried out at both low (0.25 mM) and high (2.0 mM) substrate concentrations, in view of the 2-component Michaelis-Menten model for human liver oxazaphosphorine 4-hydroxylation reported earlier (Chang et al., 1993). CYP 2C19 and some of the allelic variants of 2C9 exhibit lowKm values for CPA and/or IFA activation (Chang et al., 1993, 1997a), whereas 3A4 and 2B6 are highKm catalysts (Fig. 3). Liver microsomal CPA and IFA 4-hydroxylation activity assayed at 2 mM substrate varied over a ∼7-fold and a ∼4-fold range, respectively, in this liver panel, consistent with the large interpatient variability in pharmacokinetics seen with these anticancer prodrugs. CPA was activated at ∼2-fold higher average rate than IFA in these human liver samples, consistent with the clinical observation that an equimolar dose of IFA produces less plasma-alkylating activity than CPA in cancer patients (Sladek, 1988; Fleming, 1997).
Activation of CPA (A) and IFA (B) catalyzed by a panel of 17 human liver microsomes at 0.25 and 2 mM drug concentrations.
Data obtained for each liver sample are expressed as mean ± ½ range of duplicate determinations. Average liver activities (last set of bars at the right) were calculated as the mean activity ± S.E.M. (n = 17 human liver microsomal activities).
RSF Method for Estimation of P-450 Enzyme Contributions to CPA and IFA Activation in Human Liver Microsomes.
A substrate-activity factor-based method (RSF approach) was applied to cDNA-expressed P-450 activities with CPA and IFA as substrate to calculate microsomal P-450 activities on the basis of microsomal P-450 profiles (see Materials and Methods). The underlying premise of this approach is that the relative enzyme activity exhibited by a given cDNA-expressed P-450 form with two different P-450 substrates can be used to predict the relative activity of that same P-450 with the same pair of substrates when assayed in liver microsomes. The two substrates are respectively designated “test substrate”, whose microsomal activity is to be predicted and “diagnostic substrate”, whose microsomal activity is assayed and provides a quantitative, catalytic measure of the abundance of a specific P-450 form in liver microsomes. For example, if Supersome 2B6 is 4.9 times as active in catalyzing CPA 4-hydroxylation (test substrate) compared with 7-ethoxy-4-trifluoromethylcoumarin (7-EFC) O-deethylation (2B6 diagnostic substrate) under a particular set of assay conditions (RSF = 4.9), then the RSF method would predict that in any given HLS, the rate of CPA 4-hydroxylation catalyzed by microsomal 2B6 would be 4.9 times the 2B6-dependent microsomal 7-EFC hydroxylase activity (Code et al., 1997) assayed under the corresponding sets of assay conditions.
A panel of ten P-450 form-specific, diagnostic substrates (Table2) was used to assay the corresponding ten cDNA-expressed P-450s (both lymphoblasts and Supersomes) and a panel of 17 human liver microsomes. RSF values were calculated for each of the ten P-450s from the ratio of their CPA (or IFA) 4-hydroxylase activity to their diagnostic P-450 substrate activities (RSFCPA, RSFIFA; Table 2; see eq. 1 in Materials and Methods). The contribution of each P-450 form to CPA and IFA 4-hydroxylation in human liver microsomes was then calculated. The total predicted microsomal 4-hydroxylase activity was then determined by summing the 4-hydroxylase activities calculated for each of the ten microsomal P-450s and the percentage of total predicted microsomal 4-hydroxylase activity attributed to each P-450 form then calculated. Activities calculated by the RSF method were determined for both the lymphoblast and Supersome expression system, which gave similar results2. These are shown in Table 2 for liver sample HLS2 and are averaged over the full panel of liver microsomes in Table 3.
Contributions of different P-450 forms toward oxazaphosphorine activation in an average human liver microsome
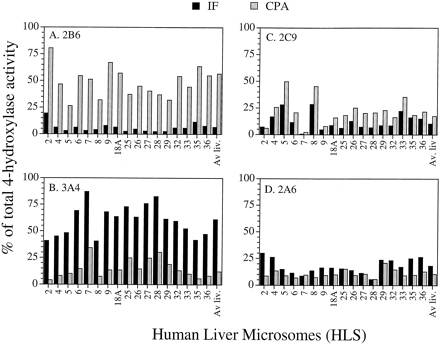
Application of the RSF method to the full panel of 17 human liver microsomes yielded the results shown in Fig.5, with the average values for the overall panel summarized in Table 3. CYP2B6 was calculated to be the major CPA 4-hydroxylase, making a contribution of 48% (based on lymphoblast expression data) or 57% of total CPA 4-hydroxylation (based on Supersome data) in an average HLS. The balance of CPA activity was contributed about equally by 2C9 (15–17%), 3A4 (12–18%), and 2A6 (10–14%). By contrast, the RSF method indicated that microsomal IFA 4-hydroxylation was primarily carried out by CYP3A4 (∼61–64% of total microsomal activity), followed by several more minor contributing enzymes: CYPs 2A6 (13–18%), 2C9 (5–10%), and 2B6 (7–11%). Although cDNA-expressed 2C19 exhibited high CPA 4-hydroxylase activity (Figs. 1 and 2), it did not contribute significantly to microsomal CPA or IFA 4-hydroxylation based on the RSF method (≤6% of the total activity). This is primarily due to the low abundance of 2C19 in human liver microsomes.
Calculated contributions of P-450s 2B6, 3A4, 2C9, and 2A6 toward overall human liver microsomal 4-hydroxylase activity determined by the RSF method.
RSF values were calculated for the panel of Supersomes for each of the substrates (CPA and IFA at 2 mM), and P-450 form-specific 4-hydroxylase activities were then determined on the basis of liver microsomal P-450 profiles. These data are based on Supersomes 3A4 and 2C9 containing cytochrome b5 (P-450:b5 ratio = 1.8), in addition to the coexpressed NADPH P-450 reductase present in all of the Supersomes. The RSF method used to calculate these data is described in detail in Materials and Methods.
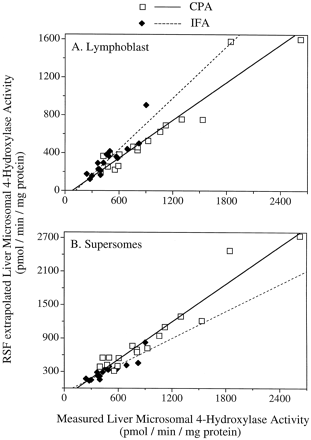
The validity of the RSF approach was supported by the excellent correlations between predicted versus measured liver microsomal 4-hydroxylation activity for CPA (r = 0.95, slope = 0.67) and IFA (r = 0.90, slope = 0.90) using lymphoblast data (Fig. 6A). Corresponding predicted versus measured values based on Supersomes data were also in excellent agreement for CPA (r = 0.96, slope = 1.13) and for IFA (r = 0.90, slope = 0.81;p < .001; Fig. 6B). In all cases,y-intercept values were close to the origin (Fig. 6). If the liver microsomal 4-hydroxylation rates had been predicted with 100% accuracy, the slopes of the correlation curves would be equal to 1.0 and the y-intercept would be equal to 0. The deviations of the slopes from 1.0 for predicted versus measured microsomal 4-hydroxylase activity may reflect the inherent variability in the measurements. Alternatively, it may suggest the existence in human liver microsomes of other contributing P-450s that have not been included in the RSF calculations. Such P-450s could include 2C18, which is an active IFA 4-hydroxylase (Figs. 1 and 2) but could not be incorporated in the RSF activity prediction due to absence of a suitable diagnostic substrate to monitor its activity in human liver microsomes.
Correlation of RSF-extrapolated versus experimentally measured total oxazaphosphorine 4-hydroxylase activity in human liver microsomes.
RSF calculations were based on the lymphoblast (A) or Supersomes expression system (B). Data are shown for CPA (open symbols) and IFA (closed symbols) in assay carried out of 2 mM drug substrate. Regression analysis yielded the curves y = 0.67x − 54.2 for CPA (r = 0.95) and y = 0.90x − 105 for IFA (r = 0.90; A), and y = 1.13x − 157 for CPA (r = 0.96) and y = 0.81x − 74.7 (r = 0.90) for IFA (B).
Effects of P-450 Form-Specific Chemical Inhibitors and Antibodies.
The P-450 form-specific contributions to oxazaphosphorine activation predicted by the RSF method were tested using P-450 form-specific chemical inhibitors to selectively inhibit microsomal activation of CPA and IFA in three individual human livers (HLS 2, 9, 36). The inhibitors used were: TAO, which is specific for P-450 3A enzymes (Chang et al., 1994; Newton et al., 1995); the CYP2B6-specific immunoinhibitory monoclonal antibody MAB-2B6 (Yang et al., 1998); and sulfaphenazole, which is a specific inhibitor of 2C9 (Newton et al., 1995; Hickman et al., 1998; Table 4). TAO (30 μM) inhibited > 90% of Supersome 3A4-catalyzed oxazaphosphorine 4-hydroxylation. TAO also inhibited to a significant extent IFA 4-hydroxylation catalyzed by human liver microsomes HLS9 (75% inhibition) and HLS36 (65% inhibition), but had a more modest inhibitory effect with HLS2 (40% inhibition). By contrast, CPA 4-hydroxylation was inhibited by TAO only ∼20% in HLS9 and <10% in HLS2 and HLS36 (Table 4). These results were confirmed with several other liver samples (52–60% inhibition of IFA 4-hydroxylase activity of HLS 25, 27, and 28; data not shown). This differential effect of TAO is consistent with the substantial contribution of 3A4 toward IFA but not CPA 4-hydroxylation determined by the RSF calculation (Fig. 5).
Inhibition of human liver microsomal oxazaphosphorine activation by P-450 form-specific inhibitors
MAB-2B6 inhibited human liver microsomal CPA 4-hydroxylation to a much greater extent than IFA 4-hydroxylation (Table 4), as predicted for 2B6 based on the RSF method (Fig. 5). Thus, MAB-2B6 inhibited CPA 4-hydroxylase in HLS2, HLS9, and HLS36 by 52%, 45%, and 33%, respectively, but had little or no effect on IFA 4-hydroxylase activity (0–13% inhibition). Lower inhibitions (∼20% of total CPA activity) were observed in liver samples where the RSF method predicted a lower 2B6 contribution (e.g., HLS27; data not shown). In control experiments, saturating amounts of MAB-2B6 (50 μg MAb-2B6/20 pmol 2B6) inhibited CPA and IFA 4-hydroxylation catalyzed by cDNA-expressed 2B6 (both lymphoblasts and Supersomes) by a maximum of 65 to 70%. MAB-2B6 also inhibited cDNA-expressed 2B6 7-EFC O-deethylase activity and human liver microsome-catalyzed, 2B6-dependent 7-EFCO-deethylation activity by 80% but was noninhibitory toward the other cDNA-expressed human CYPs (data not shown). The incomplete inhibition by MAB-2B6 of the oxazaphosphorine 4-hydroxylase activity of cDNA-expressed 2B6 may help explain our finding that MAB-2B6 inhibited liver microsomal CPA 4-hydroxylase activity to a somewhat lower extent than predicted by the RSF method (Table 4 values versus Fig. 5calculations). Finally, the 2C9 contribution toward total oxazaphosphorine 4-hydroxylase activity was determined in HLS2, HLS9, and HLS36 using 20 μM sulfaphenazole (Table 4). Inhibition by sulfaphenazole in HLS2 and HLS9 was not found to be significant, in keeping with the minor contribution (≤10%) predicted by the RSF method for these liver microsomes. Sulfaphenazole, however, caused a more significant inhibition (17–27%) of oxazaphosphorine 4-hydroxylation in microsomal sample HLS36, which confirmed the significantly higher contribution (∼20%) by 2C9 predicted by the RSF method (Fig. 5).
Immunodetection of CYP2B6 in Human Liver Microsomes.
Immunoblot analysis of the panel of 17 HLSs using MAb-2B6 revealed 2B6 protein expression in all but one liver (HLS25; Fig.7A). This confirms that 2B6 is present in a higher proportion of human liver microsomes than previously indicated using other immunochemical reagents (Shimada et al., 1994). A significant heterogeneity in 2B6 protein levels was seen among the liver samples examined, with 2B6 protein expression ranging from <1 to ∼28 pmol/mg microsomal protein. The 2B6 7-EFC O-deethylase activity component of these liver samples (Code et al., 1997) was significantly correlated with the immunoquantified levels of 2B6 in the HLSs (r = 0.615, p < .01; Fig.7B).
CYP2B6 expression in human liver microsomes.
A, Western immunoblot analysis of the panel of 17 human liver microsomes and lymphoblast and Supersome cDNA-expressed 2B6 enzymes as standards for immunoquantitation using MAB-2B6. Shown at the top are the HLS sample numbers or the pmol 2B6 standard (last six lanes). B, correlation of immunoquantitated 2B6 protein content (determined from blotting intensity obtained with MAB-2B6) and the 2B6 component of 7EFCO-deethylase activity in human liver microsomes. Immunoquantitation was performed using lymphoblast-expressed 2B6 (0.13–1.0 pmol/lane, as shown). Supersome 2B6 blotting densities were not used due to the presence of significant P-450 apoprotein in the Supersome 2B6 standard that was not accounted for in the spectrally-determined (Fe+2-CO) P-450 content of the Supersomes 2B6 samples (compare intensity of Supersome versus lymphoblast 2B6 standards at the same loading of pmol P-450). Liver HLS25 has a low but detectable 2B6 protein content (∼1 pmol 2B6/ng) that is not visible in the exposure of the Western blot presented in (B). Exclusion of sample HLS8 from the analysis (data point marked “8”) improved the correlation shown in (A) to r= 0.82 (p < .01).
Discussion
An RSF approach was used to calculate P-450 form-specific contributions to liver microsomal activation of the anticancer drugs CPA and IFA on the basis of liver microsomal P-450 profiles and cDNA-expressed enzyme activities. The validity of the RSF method was tested experimentally, enabling us to establish the relative contribution of each P-450 form to the overall rates of CPA and IFA activation in human liver. Our findings, together with earlier investigations (Chang et al., 1993; Ren et al., 1997), establish the catalytic competence of multiple human liver P-450 enzymes in the activation of CPA and IFA by the 4-hydroxylation reaction. Similar to the rat (Clarke and Waxman, 1989; Weber and Waxman, 1993), P-450 2B and 3A enzymes were found to be the principal catalysts of these 4-hydroxylation reactions. The participation of multiple human liver P-450s in oxazaphosphorine activation provides the potential opportunity to modulate the metabolism of these drugs using P-450 form-selective inhibitors and inducers. Such modulation strategies have recently been exemplified in the rat in vivo, as shown in pharmacokinetic and antitumor studies from this laboratory (Brain et al., 1998; Yu et al., 1999).
cDNA-expressed human P-450 enzymes have been utilized to study drug metabolism and to determine the principal P-450 enzyme(s) involved in the biotransformation of many individual drugs (Gonzalez and Korzekwa, 1995; Crespi and Penman, 1997). However, the application of this in vitro metabolic data to predict the specific contributions of individual P-450s to the metabolism of drugs, such as CPA and IFA, in human liver tissue, has been hampered by the absence of a well-defined framework upon which such in vitro/in vivo correlations can be drawn. Relative catalytic efficiencies exhibited by different P-450s toward CPA and IFA 4-hydroxylation in cDNA expression systems will depend not only on the intrinsic catalytic activity of each individual P-450, but also upon factors such as the relative abundance of the P-450 in human liver and the expression of accessory enzymes, such as P-450 reductase and cytochrome b5. To address these issues, we used an RSF method that utilizes the observed metabolic activity of each expressed P-450 together with measured liver P-450 profiles to calculate the catalytic contributions of each individual P-450 toward oxazaphosphorine 4-hydroxylation in a panel of human liver microsomes.
Role of CYP2B6.
Application of the RSF method to human liver microsomal CPA and IFA 4-hydroxylation indicated that 2B6 catalyzes an average of 48 to 57% of total microsomal CPA 4-hydroxylation in the liver panel under study. This observation was validated experimentally using a highly specific, inhibitory anti-2B6 monoclonal antibody, MAB-2B6 (Yang et al., 1998). CYP2B6 was characterized as a high Km/highVmax CPA 4-hydroxylase (Fig. 3) and is most likely to be a major component of the low affinity (highKm) human liver microsomal 4-hydroxylase activity described previously (Chang et al., 1993). However, given the apparent Km of 2 mM for 2B6 with CPA, the contribution of 2B6 to liver CPA activation may be lower at the plasma CPA concentrations of 0.1 to 0.7 mM typically found in the clinic (Ayash et al., 1992; Chen et al., 1995). Nevertheless, the present demonstration of the importance of 2B6 in human liver CPA activation verifies our earlier conclusions from studies using P-450 2B6-selective (but not P-450 2B6-specific) inhibitory chemical and antibody probes (Chang et al., 1993). These findings contrast with a recent study, which questioned the involvement of 2B6 in the activation of CPA in human liver, in part based on the lack of specific diagnostic probes for 2B6 (Ren et al., 1997). The P-450 form specificity of MAB-2B6 used in the present study was established by its ability to inhibit 7-EFCO-deethylation catalyzed by cDNA-expressed 2B6 by ∼80% without inhibition of any of the nine other drug-metabolizing P-450s listed in Table 2. Furthermore, Western blot analysis of human liver microsomes using MAB-2B6 detected variable amounts of 2B6 protein, which correlated well (Fig. 7) with the 2B6 component of human liver microsomal 7-EFC O-deethylase activity (Code et al., 1997). These data together with previous reports (Chang et al., 1993; Code et al., 1997; Ekins et al., 1998) establish unequivocally the presence of 2B6 in a wide range of human liver samples and the significant role that this enzyme plays in CPA bioactivation. In agreement with earlier studies (Chang et al., 1993), 2B6 does not contribute significantly toward human liver IFA 4-hydroxylation (7–11% of the total), as predicted by the RSF method (Fig. 5A; Table 3) and by the lack of significant inhibition of IFA 4-hydroxylation by MAB-2B6 (Table 4). The RSF-predicted 2B6 contribution to overall microsomal CPA activation showed a substantial interindividual variation among the human liver samples (Fig. 5A), reflecting the variability in 2B6 protein levels in the individual livers (Fig. 7). Considerable variation in 2B6 mRNA levels has also been reported in human liver (Yamano et al., 1989). This variability could be due to genetic factors, or perhaps to a differential induction of 2B6 in response to xenobiotics such as phenobarbital, dexamethasone, and rifampin, which can induce 2B6, as well as several other CPA and IFA-activating P-450s in primary human hepatocytes (Chang et al., 1997b). Exposure to these and other P-450-inducing drugs commonly used as a part of multi-drug regimen could thus be a contributing factor to the large interpatient differences reported in the clinical pharmacokinetics and biotransformation of CPA (Ayash et al., 1992; Yule et al., 1996).
Participation of CYP2C Enzymes.
That CYP2C enzymes may serve as low Kmoxazaphosphorine 4-hydroxylases in human liver microsomes was initially predicted on the basis of cDNA expression studies in HepG2 cells (Chang et al., 1993) and in a reconstituted yeast expression system containing cytochrome b5 (Chang et al., 1997a). The present study further demonstrates that cytochrome b5together with increased amounts of P-450 reductase in the Supersome expression system greatly increased the catalytic activities for CPA and IFA 4-hydroxylation for all three 2C enzymes examined (2C8, 2C9*1 and 2C19; Table 1B). However, the impact of cytochrome b5 itself on the CPA and IFA 4-hydroxylase activities of CYP2C enzymes is uncertain, given the concomitant changes in P-450 reductase levels in the Supersomes expression system. Cytochrome b5 is constitutively expressed in liver and many other mammalian tissues (Tavassoli et al., 1976) and it is unclear whether the endogenous level of cytochrome b5 is rate limiting for P-450 2C-dependent oxazaphosphorine metabolism. Individual P-450s differ in their dependence on cytochrome b5, in some cases in a manner that is substrate-dependent (Waxman and Walsh, 1983; Aoyama et al., 1990).
Among the four human CYP2C enzymes, 2C19 and some of the allelic variants of 2C9 were previously characterized as lowKm catalysts of CPA and IFA activation (Chang et al., 1997a). Because P-450 2C form-selective inhibitory antibody probes are not presently available, an attempt was made in the present study to use the RSF method to integrate cDNA expression data to predict the contribution of these enzymes to human liver microsomal CPA and IFA activation. 2C9 was thus identified as a significant enzymatic contributor in human liver, a finding that was confirmed using sulfaphenazole, which is a 2C9-selective inhibitor at low μM concentrations (Newton et al., 1995; Baldwin et al., 1995). These findings are in agreement with the contribution of 2C9 predicted by the RSF method (Fig. 5C) and are consistent with earlier clinical studies that show that sulfaphenazole can impair the elimination of CPA in some patients (Faber et al., 1975). The absence of an identified 2C18 diagnostic substrate for measurement of liver microsomal 2C18 activity precluded our application of the RSF method to determine 2C18’s catalytic contribution to liver microsomal oxazaphosphorine 4-hydroxylation. However, although 2C18 (and also 2C19) exhibited significant capacity for CPA and IFA activation [this study and (Chang et al., 1997a)], both of these P-450s are expressed at low levels in human liver, in contrast to the highly expressed 2C9 (Romkes et al., 1991). Accordingly, the catalytic contribution of 2C18 and 2C19 to liver CPA and IFA metabolism is likely to be low. CYP2C8, which is also abundant in human liver, was predicted by the RSF method to make a minor contribution to liver microsomal CPA and IFA activation, largely on the basis of its low oxazaphosphorine 4-hydroxylase activity. Finally, the 2C9 allelic variants 2C9*2 and 2C9*3demonstrated a markedly reduced capacity to activate both CPA and IFA compared with the more common 2C9*1 allele, in agreement with their reduced rates of metabolism of other substrates (Rettie et al., 1994; Crespi and Miller, 1997).
Activation of IFA by CYP3A Enzymes.
In contrast to the other P-450s, CYP3A4 demonstrated higher IFA 4-hydroxylase activity than CPA 4-hydroxylase activity in both the lymphoblast and Supersome expression systems. The predominant role of CYP3A enzymes in human liver IFA activation predicted by the RSF method (61–64% of total activity at 2 mM substrate) was verified by the significant inhibition of this microsomal activity by the CYP3A-specific inhibitor TAO (this study) and is consistent with earlier studies using anti-P-450 3A inhibitory antibodies (Chang et al., 1993; Walker et al., 1994). Supersomes 3A5 and 3A7 were also found to be catalytically competent in 4-hydroxylating both CPA and IFA, albeit somewhat less actively than 3A4, a general finding seen with several other CYP3A substrates (Aoyama et al., 1989). However, the catalytic contribution of 3A5 and 3A7 to adult human liver oxazaphosphorine activation is likely minor compared with 3A4, in view of the absence or low level of 3A5 in a majority of human livers (Shimada et al., 1994) and the preferential expression of 3A7 in fetal liver tissue (Schuetz et al., 1994). CYP3A5 may potentially play a significant role in the activation of CPA and IFA in extrahepatic tissues such as kidney, where 3A5 protein is relatively abundant (Schuetz et al., 1992). The RSF-calculated 3A4 activity does, however, also take into account the contribution of 3A5 (and probably also 3A7), insofar as testosterone 6β-hydroxylation, the CYP3A-diagnostic activity used to assess liver microsomal CYP3A levels, is catalyzed by both 3A4 and 3A5 (Aoyama et al., 1989) at a 3A4:3A5 activity ratio which is similar to the 3A4:3A5 activity ratios presented in Fig. 2 for IFA 4-hydroxylation.
Interindividual differences were noted in the extent of inhibition of IFA activation by the CYP3A-specific inhibitor TAO in a panel of 17 human liver microsomes. These differences reflect the wide range of testosterone 6β-hydroxylase activity in the liver panel (data not shown), which is indicative of the variability in CYP3A expression. 3A4 is the most abundant P-450 protein in human liver, can be induced by a broad range of drugs and xenobiotics, and accounts for an estimated 30% of total human liver P-450 content (Shimada et al., 1994). The RSF method indicated that 3A4 catalyzes a minor fraction of CPA activation in human liver (∼12–18% of the total activity), a finding that is supported by the lack of significant inhibition of this microsomal activity by TAO. These results are in sharp contrast to the observations of others (Ren et al., 1997), who concluded that P-450 3A enzymes constitute the predominant CPA 4-hydroxylases in human liver. It is evident from the present study, however, that the contribution of 3A4 toward CPA activation may vary considerably between livers, increasing from a low level of ∼5 to 10% of total activity seen in many of the livers to as high as ∼35% in case of HLS7. A considerably smaller data set (only five human livers) was used by Ren et al. (1997), raising the possibility that 2B6 (not quantitated in that study) may have been deficient or present at a very low level in that small group of samples. Indeed, substantial interindividual variation in the expression of 2B6 protein and 2B6-dependent 7EFCO-deethylase activity was seen in our panel of 17 liver microsomes (Fig. 7). The absence of data on the 3A4 expression levels in the livers investigated previously (Ren et al., 1997) makes it difficult to compare the findings in that study to those reported here.
CPA was activated at ∼2-fold higher average rate than IFA in human liver microsomes (Fig. 4), consistent with clinical data comparing the metabolism of IFA and CPA (Fleming, 1997). The lower apparent intrinsic IFA 4-hydroxylation rate may, in part, be due to the more extensive metabolism of IFA by the P-450-catalyzed N-dechloroethylation pathway, which yields therapeutically inactive, neurotoxic metabolites (Walker et al., 1994; Yu and Waxman, 1996). As noted above, all of the cDNA-expressed CYPs except for 3A4 exhibited higher 4-hydroxylase activity with CPA compared with IFA, a finding that helps explain the overall higher rates of liver microsomal CPA activation.
RSF Approach to The Calculation of Microsomal Enzyme Activities and Individual P-450 Contributions.
The frequent lack of reliable correlations between P-450-catalyzed drug metabolism data in animal models and humans (Wrighton et al., 1995;Shimada et al., 1997) has stimulated efforts to develop in vitro systems for analysis and prediction of human drug metabolism. One widely used approach involves the use of cDNA-expressed P-450 enzymes to analyze drug metabolism pathways and to identify the principal P-450 form(s) responsible for the metabolism of a drug substrate (Crespi and Penman, 1997). P-450 expression systems offer an unlimited enzyme supply and a high consistency of enzyme preparations, but suffer from limitations associated with the multiplicity of drug-metabolizing human P-450 enzymes, the relatively low levels of expression that can be achieved in stable mammalian cell culture, and the lack of a framework for establishing the relative role of a P-450 in a P-450-catalyzed metabolic reaction in vivo. In cases where multiple P-450 enzymes contribute to the metabolism of a specific drug, it is not only important to identify those P-450s that are catalytically active, but also to determine the relative abundance of each P-450 enzyme in the tissue of interest (e.g., human liver), so that the principal P-450s active in the tissue in vivo may be predicted. Rigorous quantitation of the levels of all P-450 forms expressed in human liver is not currently feasible, because diagnostic reactions and specific inhibitory probes and antibodies are unavailable in several cases. Such in vitro/in vivo correlations are further complicated by the occurrence of allelic polymorphisms that impact on enzymatic activity and by the presence in individual livers of varying levels of P-450 reductase and cytochrome b5, which in turn, may be different from the levels found in cDNA-expressed P-450 preparations. Consequently, enzyme turnover numbers may vary substantially between in vitro systems, making an accurate prediction of the balance of P-450 enzyme contributions in human liver tissue difficult.
The RSF-based approach presented here addresses many of these limitations and provides a conceptually and mathematically simple way to integrate data obtained with cDNA-expressed P-450s to predict the balance of contributing P-450 enzymes in human liver. The net result of calculations using the present RSF approach is identical with that of the RAF approach described earlier (Crespi, 1995). The principal difference is that the RSF approach is based on the ratio of the activity of the cDNA-expressed preparation toward the test and diagnostic substrates, whereas the RAF approach is based on the ratio of activity of the cDNA-expressed and human liver microsomal enzymes to each other. Specific activity data and relative enzyme contributions predicted by both calculation methods are refractory to differences in absolute turnover number, allowing one to combine data from P-450 cDNA expression systems that differ in their levels of P-450 reductase and cytochrome b5. The RSF-predicted P-450 form-specific contributions to human liver microsomal CPA and IFA activation presented here were in good agreement with the inhibition data determined experimentally with P-450 form-specific inhibitors (Table 4). Furthermore, the RSF-based method for estimation of human liver microsomal oxazaphosphorine 4-hydroxylase activity showed strong correlations with the measured 4-hydroxylation rates, independent of which expression panel was used (Fig. 6). A perfect correlation of predicted with measured enzyme activity (i.e., slope of the correlation line equals 1.0 and line passes through the origin, cf., Fig. 6) was not observed and can only be obtained when all of the active P-450s for the particular reaction under investigation are accounted for and are included in the RSF calculation. The deviations from a perfect correlation in the present case may be due to other active CPA or IFA 4-hydroxylase P-450s that are unidentified or were not included in the RSF calculation due to the unavailability of a suitable diagnostic microsomal substrate (e.g., 2C18).
Assumptions and Limitations of RSF Method.
Although the RSF method was found to be good at predicting liver microsomal activities and P-450 enzyme contributions, its application is premised on the following assumptions: 1) The enzyme activity values determined with the diagnostic and test substrates must be a linear function of the amounts of enzyme present in each system (This ensures linearity of the comparison across the range of activities being compared. There is no requirement, however, that the diagnostic or test substrates be assayed at saturating concentrations.); 2) The diagnostic substrate must be truly P-450 form-selective under the conditions of the assay and in the tissue under study (This ensures that the diagnostic activity values, as measured in human liver microsomes, accurately reflect the abundance of the specific P-450 of interest. If the diagnostic substrate activity encompasses more than one P-450 form [as in the case of testosterone 6β-hydroxylase activity in human liver tissue, which measures total CYP3A activity without distinguishing 3A4 from 3A5 or 3A7 activity], then the RSF method can only determine the total contribution of those enzymes, without separation of the contributions made by each individual P-450 form. As a practical matter, an overall selectivity of 80% or higher should be adequate for this approach.); 3) The day-to-day variability in the enzyme assay values should be small (The assays used in the present study demonstrate day-to-day variabilities of up to 10 to 20%. The use of reference lots of enzyme with known levels of activity is recommended to provide a measure of assay performance on each day.); and 4) The RSF approach assumes that any factors that influence the rate of metabolism of the diagnostic substrate do so equally for the test substrate, such that the ratio of activities of the test and diagnostic substrates is constant for each enzyme preparation. An example of this effect is the differential impact that cytochrome b5 can have on a P-450’s activity toward different substrates. Our finding that RSF values for some of the CPA or IFA 4-hydroxylating P-450s differed by ∼2-fold when determined in the lymphoblast versus Supersome expression system (Table 2) indicates, however, that this assumption may not be fully met under all circumstances. Thus, the ratio of test to diagnostic substrate activity may not necessarily be intrinsic to the P-450 alone and may be influenced by factors specific to each expression system. An example of this was seen for 2B6 activity, which was highly stimulated by cytochrome b5 with the diagnostic substrate 7-EFC, but not with the test substrates CPA and IFA. By contrast, an example where assumption (4) has been validated is provided by the comparisons of (S)-warfarin 7-hydroxylase and diclofenac 4′-hydroxylase activities catalyzed by CYP2C9*1 in lymphoblastoid cells, baculovirus-insect cell systems and human liver microsomes (Crespi and Miller, 1997).
In summary, an RSF method was developed and shown to be useful in predicting the contribution of individual P-450 enzymes to the activation of the anticancer prodrugs CPA and IFA, as verified experimentally using P-450 form-specific chemical and antibody inhibitors. P-450 2B6 was identified as the major enzyme responsible for CPA 4-hydroxylation, whereas P-450 3A4 was confirmed to be the principal catalyst of IFA 4-hydroxylation. CYPs 2A6 and 2C9 were both found to play more minor roles in the activation of CPA and IFA in human liver microsomes. The catalytically active oxazaphosphorine 4-hydroxylase P-450s that are of low abundance in human liver, such as CYPs 2C18 and 2C19, may potentially find use in an intratumoral expression cancer gene therapy/chemotherapy strategy (Chen and Waxman, 1995; Waxman et al., 1999) to elevate the therapeutic efficacy of these otherwise toxic alkylating agents. Finally, the RSF method may find widespread use both in predicting potential drug-drug interactions and in predicting metabolism by human liver tissue of new drug substrates based on in vitro assays using lymphoblasts, Supersomes, or other panels of cDNA-expressed human P-450 enzymes.
Footnotes
-
Send reprint requests to: Dr. David J. Waxman, Department of Biology, Boston University, 5 Cummington St., Boston, MA 02215. E-mail: djw{at}bio.bu.edu
-
This work was supported in part by National Institutes of Health Grant CA49248 (D.J.W.).
-
↵2 RSF calculations based on the Supersomes expression system were based on activities determined in the presence of cytochrome b5, with the exception of 2B6, where Supersomes minus cytochrome b5 data was used. 2B6 + cytochrome b5 Supersomes showed ∼5-fold higher 2B6 diagnostic substrate activity (7-EFC O-deethylation) compared with 2B6 without cytochrome b5, whereas CPA 4-hydroxylase activity was only ∼35% higher with cytochrome b5, indicating that there is a significant substrate dependence to the cytochrome b5 stimulation of 2B6. This differential stimulation of test versus diagnostic substrate activity significantly alters the results of RSF-based calculations and is recognized as a limitation of this method (SeeDiscussion).
- Abbreviations used are::
- CYP
- cytochrome P-450
- CPA
- cyclophosphamide
- IFA
- ifosfamide
- HLS
- human liver microsomal sample
- 7-EFC
- 7-ethoxy-4-trifluoromethylcoumarin
- RSF
- relative substrate-activity factor
- RAF
- relative activity factor, TAO, troleandomycin
- 2C9*1
- 2C9-Arg144 allele
- 2C9*2
- 2C9-Cys144 allele,2C9*3, 2C9-Leu359 allele
- MAB-2B6
- monoclonal antibody specific to 2B6
- Received December 23, 1998.
- Accepted March 18, 1999.
- The American Society for Pharmacology and Experimental Therapeutics