Abstract
Carboxylesterases play important roles in the metabolism of endogenous and foreign compounds, therefore, xenobiotic regulation of carboxylesterase gene expression has both physiological and pharmacological significance. We previously reported that liver microsomal esterase activity was significantly decreased in rats treated with dexamethasone accompanied by a decrease in immunoreactive proteins of rat hydrolase A, B, and C. The aim of this study was to determine whether the suppressed expression of these enzymes was linked to the change of the mRNA levels, and whether cultured hepatocytes responded similar to whole animals to this chemical. Northern blotting analyses demonstrated that the levels of the corresponding mRNA were markedly decreased in rats treated with dexamethasone, suggesting that the suppressed expression is achieved throughtrans-suppression and/or increased degradation of the transcripts. Exposure of cultured rat hepatocytes to nanomolar levels of dexamethasone markedly decreased the levels of immunoreactive proteins of hydrolase A, B, and C. In contrast, exposure of cultured human hepatocytes to dexamethasone caused a slight increase in HCE-1 and HCE-2, two major forms of human liver microsomal carboxylesterases. The inductive effects in human hepatocytes were observed only when micromolar concentrations of dexamethasone were used. These results suggest that a major species difference exists regarding the regulation of carboxylesterase gene expression by dexamethasone. Both the glucocorticoid receptor and the pregnane X receptor are known to mediate dexamethasone action. Differential concentrations required suggest that suppression of rat hydrolases is mediated by the glucocorticoid receptor, whereas the induction of human carboxylesterases is mediated by the pregnane X receptor.
Carboxylesterases play an important role in the metabolism of endogenous lipids and foreign compounds, such as drugs and pesticides (Junge and Krisch, 1975; Heymann, 1982; Parkinson, 1995; Satoh and Hosokawa, 1998). In addition to hydrolyzing numerous chemicals, carboxylesterases can catalyze transesterification reaction, which accounts for the conversion of cocaine (a methyl ester) to ethylcocaine (the corresponding ethyl ester) in the presence of ethyl alcohol (Boyer and Petersen, 1992). Carboxylesterase activity is widely distributed in mammalian tissues, with the highest levels present in liver microsomes (Parkinson, 1995; Satoh and Hosokawa, 1998). High abundance of carboxylesterases in the liver is linked to certain cellular structural roles, particularly in directing protein targeting (Medda et al., 1987;Ovnic et al., 1991). For example, egasyn, a liver microsomal carboxylesterase identified in rat and mouse, binds to β-glucuronidase via its active site, which results in sequestration of this enzyme within the endoplasmic reticulum (Medda et al., 1987;Ovnic et al., 1991). In rabbits, microsomal carboxylesterases are shown to interact with and regulate the secretion of acute-phase-response proteins, such as C-reactive protein (Macintyre et al., 1994).
Multiple forms of carboxylesterases are identified in several mammalian species (Robbi and Beaufay, 1994; Satoh and Hosokawa, 1998). We previously reported the isolation of cDNAs encoding four rat and three human carboxylesterases. The rat enzymes are designated hydrolase A, B, C, and S (Yan et al., 1994; 1995a,b,c), whereas the human enzymes are designated PCE-1, PCE-2, and PCE-3, respectively (Yan et al., 1999). Hydrolase A, B, C, and S are ∼70% identical at both the nucleotide and the derived amino acid sequences, with the exception of hydrolase B and C, which are ∼95% identical. PCE-1 and PCE-2, originally cloned from the placenta, are highly identical with liver HCE-1 and HCE-2, respectively (Kroetz et al., 1993; Pindel et al., 1997). PCE-3 has an open reading frame only for 268 amino acids, half the size of a regular carboxylesterase (Yan et al., 1999).
A wide range of drugs and other xenobiotics are found to alter the expression of carboxylesterases in animals, but the inducibility is minimal in most species compared with cytochrome P-450 (CYP)3 enzymes (Morgan et al., 1994a,b; Parkinson, 1995; Satoh and Hosokawa, 1998). In rats, phenobarbital and clofibrate, two potent CYP inducers, cause a small increase (15–30%) of activity in hydrolyzingpara-nitrophenylacetate (Morgan et al., 1994a). The mechanism on the lack of inducibility is unclear, probably due to the fact that carboxylesterase genes are constitutively expressed at a relatively high level (Morgan et al., 1994a,b). In contrast to induction, suppression of carboxylesterase expression is profound by many chemicals (Morgan et al., 1994a,b; Watson et al., 1994; Satoh and Hosokawa, 1998). Treatment of mature rats with dexamethasone and β-naphthoflavone causes as much as an 80% decrease in hydrolytic activity toward para-nitrophenylacetate and the corresponding immunoreactive proteins of hydrolase A, B, C, and S (Morgan et al., 1994a; Yan et al., 1995b).
The aim of this study was to determine whether the suppressed expression of these rat enzymes was linked to the change of the mRNA levels, and whether cultured hepatocytes responded similar to whole animals to dexamethasone. Northern blotting analyses demonstrated that the levels of the corresponding mRNA were markedly decreased in rats treated with dexamethasone, suggesting that the suppressed expression is achieved through trans-suppression and/or increased degradation of the transcripts. In cultured rat hepatocytes, exposure to dexamethasone resulted in a decrease of hydrolase A, B, and C. In a striking contrast, exposure of cultured human hepatocytes to dexamethasone caused a slight increase in HCE-1 and HCE-2, two major forms of human liver microsomal carboxylesterases. These results suggest that a major species difference exists regarding the regulation of carboxylesterase gene expression by dexamethasone.
Materials and Methods
Chemicals and Supplies.
Dexamethasone, rifampicin, and 3-methylcholanthrene were obtained from Sigma Chemical Co. (St. Louis, MO). The goat anti-rabbit IgG conjugated with alkaline phosphatase was purchased from Pierce (Rockford, IL). The isothermal DNA sequencing kit was purchased from Epicentre Technology Inc. (Cleveland, OH). Sprague-Dawley rats (8–10 week old) were purchased from Charles River (Wilmington, MA). Cell culture media, human liver cDNA library, and the PLATINUM Taq DNA polymerase were purchased from Life Technologies (Gaithersburg, MD). Anti-rat CYP3A23/2 antibody was purchased from the XenoTech LLC (Kansas City, KS). Unless otherwise indicated, all other reagents were purchased from Fisher Scientific (Pittsburgh, PA).
Heterologous Expression of HCE-1, HCE-2, and CYP3A4.
We previously isolated from human placenta several cDNAs encoding three distinct proteins: PCE-1, PCE-2, and PCE-3 (Yan et al., 1999). PCE-1 is identical with human liver HCE-1, whereas PCE-2 is 99% identical with human liver HCE-2. This study primarily used liver microsomal samples, therefore, the names of HCE-1 and HCE-2 are used thereafter. To prepare plasmid constructs for stable transfection, cDNAs encoding PCE-1 and PCE-2 were released from the corresponding original plasmids withKpnI and XbaI endonuclease digestion. The released cDNA fragments were subcloned into the pIZ/V5-His plasmid, an insect cell expression vector (Invitrogen, Carlsbad, CA). The construct for CYP3A4 was prepared by polymerase chain reaction (PCR), essentially as described previously (Yan et al., 1995c). The sense primer (5′-TCAggtaccATGGCTCTCATCCCAGAC-3′) was extended to include aKpnI site, whereas the antisense primer (5′-GACtctagaGGTCTCTGGTGTTCTCAG-3′) was extended to include aXbaI site (Yan et al., 1995c). The template DNA for PCR amplification was prepared from a human liver cDNA library as described previously (Yan et al., 1995c; 1999). The PCRs were conducted with the PLATINUM Taq high fidelity polymerase for a total of 30 cycles. The PCR products were extracted with phenol-chloroform and precipitated with ethanol, followed by KpnI andXbaI digestion and agarose gel electrophoresis purification. The endonucleases-treated fragments were inserted into the pIZ/V5-His plasmid. All plasmid constructs were subjected to sequencing analysis. For stable transfection, Sf9 cells at the log phase were washed twice with Grace's Insect media to remove any trace of serum and transfected with Insectin-Plus liposomes (Invitrogen). Transfection was terminated by adding an equal amount of Trichoplusia ni Medium-Formulation Hink (TNM-FH) medium, and the transfected cells were incubated at 27°C for 48 h. Cells were split and allowed to attach for 15 min in TNM-FH medium, and then the medium was replaced with selective medium (TNM-FH plus Zeocin at 350 μg/ml). The stable transfectants were obtained by growing under selective conditions for ∼3 weeks with a change of the medium every 4 days.
Antibody Preparation.
Anti-peptide antibodies were raised in New Zealand White rabbits. The sequences of the peptides were: HCE-1, H2N-CEKPPQTEHIEL-COOH; HCE-2, H2N-CQELEEPEERHTEL-COOH; and CYP3A4, H2N-CVKRMKESRLEDTQKHRVDFLQ-COOH. Peptides were synthesized and conjugated with keyhole limpet hemocyanin (Genemed Synthesis Inc., South San Francisco, CA). The first immunization was conducted by injecting each rabbit s.c. on the back with 50 μg of the conjugated peptide emulsified with equal volume of Freund's complete adjuvant (Pierce). The second immunization was conducted 4 weeks later, at which time the conjugated peptide was emulsified with an equal volume of Freund's incomplete adjuvant. Final boosting was conducted 4 weeks later by giving i.v. injection (30 μg in 200 μl of saline per rabbit). Antiserum was collected 1 week after the final immunization.
The antibody specific to the peptide was purified by immunoaffinity chromatography. The peptide lacking the conjugated keyhole limpet hemocyanin was covalently bound to SulfoLink gel (Pierce) via its N-terminal cysteine residue at a ratio of 100 μg peptide over 1 ml of gel according to the manufacturer. The SulfoLink gel was immobilized by iodoacetyl moiety on the surface, which was highly reactive to sulfhydryls. Immunoaffinity purification was conducted, essentially as described by Harlow and Lane (1988). The antisera (2 ml) were diluted 10-fold in 10 mM Tris-HCl (pH 7.5), and loaded to the peptide SulfoLink gel column (10 ml). The antibody solution was reapplied to the column an additional two times to ensure complete binding. The column was then washed with 20 bed-volumes of 10 mM Tris-HCl (pH 7.5), and then 20 bed-volumes of 500 mM NaCl, 10 mM Tris (pH 7.5). The antibody specific to the peptide was then eluted by 100 mM glycine (pH 2.5), and the eluate (containing anti-peptide) was collected in tubes containing 1 bed-volume of 1 M Tris-HCl (pH 8.0). The eluate was dialyzed against PBS containing 0.02% sodium azide at 4°C overnight, aliquoted, and stored at −20°C.
Hepatocyte Culture and Chemical Treatment.
Rat liver was perfused with collagenase-containing buffer through the portal vein, and hepatocytes were prepared as previously (Sidhu et al., 1993; Yan et al., 1995b). Human hepatocytes were isolated from human liver tissue obtained as surgical waste or from rejected donor livers by a modified two-step collagenase digestion method (LeCluyse et al., 1996; Strom et al., 1997). Briefly, human liver tissue (25–100 g) was encapsulated and perfused with calcium-free buffer containing 5.5 mM glucose and 0.5 mM EGTA for 10 to 15 min at a flow rate of 30 to 50 ml/min followed by perfusion with buffer containing 1.5 mM calcium and collagenase (0.3–0.4 mg/ml) for 20 to 25 min. Hepatocytes were dispersed from the digested liver in Dulbecco's modified Eagle's medium (DMEM) containing 5% fetal calf serum, insulin, and dexamethasone and washed by low-speed centrifugation (70g, 4 min). Cell pellets were resuspended in supplemented DMEM and 90% isotonic Percoll (3:1 v/v). Cell suspensions were centrifuged at 100g for 5 min. The resulting pellets were resuspended in fresh medium and washed once by low-speed centrifugation. Hepatocytes were resuspended in supplemented DMEM and viability was determined by trypan blue exclusion. Hepatocyte viability at isolation was >90% for rats and 80 to 90% for humans, respectively. Hepatocytes were plated onto collagen-coated Permanox culture dishes at a density of 4 to 4.5 million hepatocytes per 60-mm dish. The plated hepatocytes were allowed to attach for 2 to 4 h at 37°C in a humidified chamber with 95%/5% air/CO2. Culture dishes were gently swirled and medium containing unattached cells was then aspirated. Fresh ice-cold modified Chee's medium containing 6.25 mg/ml insulin, 6.25 mg/ml transferrin, 6.25 ng/ml selenium, and 0.25 mg/ml Matrigel was added to each dish and cultures were returned to the humidified chamber. Medium was changed on a daily basis thereafter. Unless otherwise specified, hepatocytes were maintained for 48 to 72 h before initiating treatment with xenobiotics. Groups of hepatocyte cultures (n = 3 dishes per treatment group) were then treated for 3 consecutive days with drug at various concentrations. Control cultures were treated with vehicle alone (0.1% dimethyl sulfoxide). At the end of each study period, cells were harvested for the preparation of microsomes as described previously (Morgan et al., 1994a). Use of human tissues was approved by the University Institutional Review Committee.
Animal Treatment.
Sprague-Dawley rats (five per group) were treated as described previously (Yan et al., 1995b). All rats were housed in an American Association for Accreditation of Laboratory Animal Care. Mature male rats were injected i.p. once daily for four consecutive days with dexamethasone (50 mg/kg). Livers were harvested 24 h after the last injection. Total RNA from control or xenobiotic-treated Sprague-Dawley adult rats was isolated with a TRI Reagent RNA extraction solution according to the instruction by the manufacturer. RNA samples from each group of rats were pooled and the concentrations were determined from the absorbance at 260 nm, with 1 A unit equal to 40 μg/ml. Liver microsomes were prepared by differential centrifugation and stored −80°C as suspension in 250 mM sucrose as described previously (Morgan et al., 1994a).
Other Assays.
Proteins were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) described elsewhere (Morgan et al., 1994a). Western immunoblotting with antibodies against carboxylesterases and CYP3A4 as described previously (Yan et al., 1995b). The staining intensity was determined with a laser scanning densitometer (Biomed Instruments, Inc., Fullerton, CA). Northern blotting with cDNAs encoding hydrolase A, B, or S was conducted as described previously (Yan et al., 1994,1995a,b).
Results
Antibody Specificity.
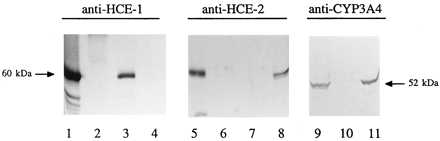
Regulation of carboxylesterase gene expression in humans and rats was studied primarily by immunochemical techniques. Anti-peptide antibodies were raised in rabbits and subjected to affinity chromatography. The specificity of these antibodies was established with the corresponding recombinant proteins. As shown in Fig. 1, in all cases, the antibody against a peptide derived from HCE-1, HCE-2, or CYP3A4 detected a single protein only in the Sf9 cells expressing the corresponding recombinant protein. Similarly, a major protein was detected by each antibody in human liver microsomes, and the microsomal protein comigrated with the corresponding recombinant protein (Fig. 1). As expected, HCE-1 and HCE-2 had a similar molecular mass (∼60 kDa), whereas CYP3A4 was a 52-kDa protein. It is known that carboxylesterases undergo glycosylation, which results in an increase in the molecular mass (Kroetz et al., 1993; Yan et al., 1995b). Comigration of the recombinant proteins with their native counterparts (liver microsomal protein) suggests that the insect cell expression system biosynthesizes these enzymes similar to hepatocytes.
Specificity of antibodies against peptides derived from HCE-1, HCE-2, or CYP3A4.
Human microsomes (5 μg) or Sf9 cell lysates (5 μg) were subjected to SDS-PAGE as described in Materials and Methods. The cell lysates were prepared from either control or transfected Sf9 cells. Samples were transferred electrophoretically to a Trans-Blot nitrocellulose membrane. The immunoblots were blocked in 5% nonfat dry milk and then incubated with the antibody (10 μg/ml) against HCE-1 (left), HCE-2 (middle), or CYP3A4 (right). The primary antibody was then located by alkaline phosphatase-conjugated goat anti-rabbit IgG. Lanes 1, 5, and 9 contained human liver microsomes; lanes 2, 6, and 10 contained lysates from nontransfected Sf9 cells; lanes 3 and 7 contained lysates from HCE-1-transfected Sf9 cells; lanes 4 and 8 contained lysates from HCE-2-transfected Sf9 cells; and lane 11 contained lysates from CYP3A4-transfected Sf9 cells.
Dexamethasone Suppresses Expression of Hydrolase A, B, C, and S in Rats and Cultured Rat Hepatocytes.
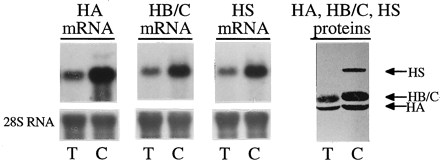
We previously reported that hydrolytic activity ofpara-nitrophenylacetate in liver microsomes was significantly decreased in rats treated with dexamethasone (Morgan et al., 1994a). We also reported that the treated rats expressed lower levels of hydrolase A, B, C, and S as determined by immunoblotting (Morgan et al., 1994a; Yan et al., 1995b). To determine whether the suppressed expression was linked to the change of the corresponding mRNA, male mature rats were treated with dexamethasone and total RNA was isolated as described previously (Yan et al., 1995b). The RNA samples were then subjected to agarose gel electrophoresis and detected with probes prepared from cDNA encoding hydrolase A, B, or S. Microsomes from these livers were also prepared and analyzed by immunoblotting with anti-hydrolase S. This antibody has been shown to recognize hydrolase A, B, C, and S (Yan et al., 1995b). Hydrolase B and C are ∼95% identical and electrophoretically indistinguishable under the conditions used (Yan et al., 1995c). As shown in Fig.2, the dosing regimen of dexamethasone markedly decreased the levels of mRNA encoding hydrolase A, B, C, or S. This decrease was accompanied by a decrease in the corresponding immunoreactive proteins (∼50%).
Effect of dexamethasone on the expression of hydrolase A, B/C, and S; and CYP3A23 in rats.
Sprague-Dawley male rats (10 weeks old, five per group) were injected i.p. once with dexamethasone (50 mg/kg) for 4 consecutive days. Total RNA was isolated from treated (T) or control (C) rats with a TRI Reagent as described by the manufacturer. Pooled total RNA samples (10 μg) were subjected to formaldehyde agarose electrophoresis and transferred to a Nytran nylon membrane by a vacuum blotting system. The blots were detected with 32P-labeled probes from cDNA encoding hydrolase A, B, or S (left). Immunoblotting analyses for the expression of these carboxylesterases were conducted with anti-hydrolase S, which has been shown to recognize hydrolase A (HA), hydrolase B (HB), hydrolase C (HC), and hydrolase S (HS). To show the change of microsomal hydrolase S, 25 μg of microsomal proteins from pooled samples were used.
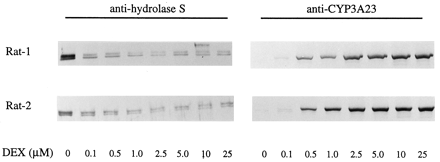
Next we examined whether dexamethasone suppressed the expression of these carboxylesterases in cultured rat hepatocytes. Rat hepatocytes were prepared and treated with dexamethasone as described inMaterials and Methods. Expression of carboxylesterases in the microsomes from these cells was determined by immunoblotting. As shown in Fig. 3, exposure of rat hepatocytes to 0.1 micromolar dexamethasone caused a marked decrease (∼50%) in the immunoreactive proteins of hydrolase A, B, and C. Such suppressive effects exhibited a concentration-dependent manner and reached the maximum when 2.5 micromolar dexamethasone was used. Hydrolase S is a secretory enzyme, therefore, its presence in the microsomes was not detectable under the conditions used. In contrast, the CYP3A23 gene was drastically induced, but such inductive effects were not observed when 0.1 micromolar dexamethasone was used. Dexamethasone at higher concentrations (2.5–25 μM), which showed no additional suppressive effect on carboxylesterase expression, markedly elevated the levels of CYP3A23 (Fig. 3). These results suggest that the pathway responsible for regulating carboxylesterase genes is more sensitive than that for CYP3A genes in rats, and cultured hepatocytes responds similar to whole animals to dexamethasone-mediated regulation in the expression of carboxylesterases and CYP3A enzymes.
Effect of dexamethasone on the expression of hydrolase A, B/C, and S; and CYP3A23 in cultured rat hepatocytes.
Rat hepatocytes were cultured in supplemented DMEM for 48 h and then exposed to dexamethasone at various concentration (0.1–250 μM) for an additional 3 days with a daily change of the culture media. Hepatocytes were harvested 24 h after the last treatment, and microsomes were prepared from both control and treated hepatocytes. Microsomal samples (5 μg) were pooled from three individual culture dishes and subjected to SDS-PAGE. Expression of hydrolase A, B, and C, as well as CYP3A23/2 (2 μg) was determined by immunoblotting as described in Fig. 1. Rat hydrolases display as two major bands: the top band represents hydrolase B and C, whereas the bottom one represents hydrolase A. Hydrolase S was undetected in the microsomes under the conditions used. DEX, dexamethasone.
Dexamethasone Induces Expression of Carboxylesterase Genes in Human Hepatocytes.
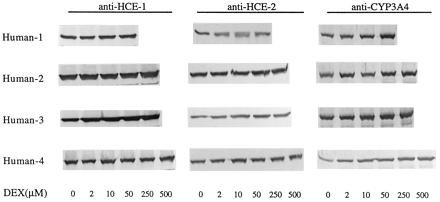
We next examined the effect of dexamethasone on the expression of carboxylesterase genes in cultured human hepatocytes. Human liver tissues obtained as surgical waste or from rejected donor livers were encapsulated and perfused by a two-step collagenase digestion method (LeCluyse et al., 1996; Strom et al., 1997). Similar to rat hepatocytes, human hepatocytes were exposed to dexamethasone at various concentrations for 3 consecutive days, and microsomes were prepared. As shown in Fig. 4, microsomes from all individuals contained both carboxylesterases. In contrast to the suppression of rat carboxylesterase expression, human HCE-1 and HCE-2 were moderately induced (∼20%). The inductive effects exhibited a concentration-dependent manner but were observed only when concentrations were 10 micromolars or higher (Fig. 4). A similar dependence on the concentrations was observed with CYP3A4 induction in these samples. Inductive effects in human hepatocytes and suppressive effects in rat hepatocytes suggest that a major species difference exists regarding the dexamethasone-mediated regulation in the expression of these carboxylesterase genes (Fig. 4).
Effect of dexamethasone on the expression of HCE-1, HCE-2, and CYP3A4 in cultured human hepatocytes.
Human hepatocytes were isolated from human liver tissues obtained as surgical waste or from rejected donor livers by a two-step collagenase digestion method. Hepatocytes were cultured in supplemented DMEM for 72 h and then exposed to dexamethasone at various concentrations (0.1–500 μM) for an additional 3 days with a daily change of the culture media. Hepatocytes were harvested 24 h after the last treatment, and microsomes were prepared from both control and treated hepatocytes. Microsomal samples (5 μg) were pooled from three individual culture dishes and subjected to SDS-PAGE. Expression of hydrolase HCE-1, HCE-2, and CYP3A4 was determined by immunoblotting as described in Fig. 1.
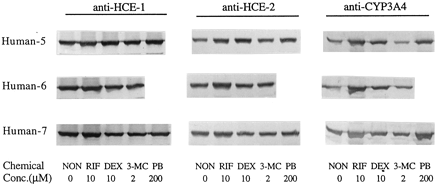
These studies were extended to include other CYP inducers on the expression of human carboxylesterases. These chemicals included rifampicin (10 μM), phenobarbital (200 μM), and 3-methylcholanthrene (2 μM). As shown in Fig.5, both rifampicin and phenobarbital caused a moderate induction of HCE-1 and HCE-2 with rifampicin being more potent, and 3-methylcholanthrene slightly suppressed the expression of both enzymes. A similar induction profile of CYP3A4 was observed among these chemicals, suggesting that the same signaling pathway is involved in the induction of both human carboxylesterase and CYP3A4.
Effect of rifampicin, dexamethasone, phenobarbital, and 3-methylcholanthrene on the expression of HCE-1, HCE-2, and CYP3A4 in cultured human hepatocytes.
Human hepatocytes were isolated from human liver tissues obtained as surgical waste or from rejected donor livers by a two-step collagenase digestion method. Hepatocytes were cultured in supplemented DMEM for 72 h and then exposed to rifampicin (10 μM), dexamethasone (10 μM), phenobarbital (200 μM), and 3-methylcholanthrene (2 μM) for 3 days with a daily change of the culture media. Microsomal samples (5 μg) were pooled from three individual culture dishes and subjected to SDS-PAGE. Expression of hydrolase HCE-1, HCE-2, and CYP3A4 was determined by immunoblotting as described in Fig. 1. RIF, rifampicin; DEX, dexamethasone; 3-MC, 3-methylcholanthrene; and PB, phenobarbital. ∗, hepatocytes from this person were treated with 50 μM dexamethasone.
Discussion
Dexamethasone is a synthetic steroid and is widely used for therapeutic purposes (Garland et al., 1999; Philip et al., 1999;Verhoef et al., 1999). This drug is known to modulate the expression of several important drug-metabolizing enzymes, which contributes significantly to drug-drug interactions (Junge and Krisch, 1975;Heymann, 1982; Parkinson, 1995; Satoh and Hosokawa, 1998). We have reported that liver microsomal esterase activity is significantly decreased in rats treated with dexamethasone (Morgan et al., 1994a). In this report, we have extended these studies to include cultured rat and human hepatocytes. Exposure of rat hepatocytes to nanomolar levels of dexamethasone results in a marked decrease of hydrolase A, B, and C. In contrast, exposure of cultured human hepatocytes to dexamethasone causes a slight increase in HCE-1 and HCE-2, two major forms of human liver microsomal carboxylesterases (Kroetz et al., 1993; Pindel et al., 1997). The inductive effects are observed only when higher levels of dexamethasone are used. These results suggest that a major species difference exists regarding the regulation of carboxylesterase gene expression by dexamethasone.
Dexamethasone-mediated regulation of carboxylesterase genes also exhibits an isoform-specific manner. We describe here that exposure to dexamethasone causes a marked decrease of hydrolase A, B, C, and S in both rats and cultured rat hepatocytes (Figs. 2 and 3). However, previous studies with nondenaturing gel electrophoresis for staining esterase activity have identified a dexamethasone-induced carboxylesterase (Morgan et al., 1994b). The induced enzyme is electrophoretically distinct from hydrolase A, B, C, and S. Hosokawa et al. (1993) used specific substrates to study the regulation of dexamethasone on carboxylesterase gene expression in rats. Hydrolytic activity toward para-nitrophenylacetate and butanilicaine is significantly decreased, suggesting that hydrolases A, B, and C collectively contribute to the hydrolysis of both compounds. Hydrolase S is a secretory carboxylesterase, therefore, it contributes little to the overall hydrolytic activity in the liver microsomes (Yan et al., 1995b). In contrast, the hydrolytic activity toward isocarboxazid is significantly increased in the dexamethasone-treated rats. The isocarboxazid hydrolase(s) is also induced by pregnenolone-16α-carbonitrile, an antagonist of glucocorticoid steroid (Hosokawa et al., 1993). The differential responses of carboxylesterase isoforms suggest that these hydrolytic enzymes have physiological significance and suppression of all isoforms has a detrimental effect on normal cellular function. Coregulation of hydrolase A, B, C, and S as well as HCE-1 and HCE-2 suggests that these carboxylesterase genes evolved from the same ancestral gene and contain the same or a similar cis-response element mediating dexamethasone action.
The DNA response elements in the carboxylesterase genes determine the induction or suppression by a xenobiotic, but the type of CYP inducers may provide little relevance. Expression of rat hydrolase S, for example, is significantly suppressed by isoniazid but slightly increased by streptozotocin; both of them are 2E1 inducers (Yan et al., 1995b). The CYP1A enzyme inducers, β-naphthoflavone and 3-methylcholanthrene, have opposing effects; the former compound suppresses the expression of hydrolase S, whereas the latter compound induces it. Expression of hydrolase S is suppressed by perfluorodecanoic acid but unaffected by clofibric acid; both of them are CYP4A inducers. Structurally related compounds may also have different effects on carboxylesterase expression. Dexamethasone and pregnenolone 16α-carbonitrile are both synthetic steroids and both induce CYP3A gene expression. However, only dexamethasone suppresses the expression of rat RL1 and RH1 (e.g., hydrolase A, B), but the other has little effect on both enzymes (Hosokawa et al., 1993). The differential response of carboxylesterase genes to the same type of CYP inducers suggests that these chemicals use multiple pathways to exert their biological effects.
Regulation of carboxylesterase gene expression by dexamethasone is likely due to the alteration of transcriptional rate and/or mRNA stability inasmuch as the decreased levels of immunoreactive proteins are accompanied by decreased levels of the corresponding mRNA (Fig. 2). Two types of nuclear receptors are known to mediate dexamethasone action: the glucocorticoid receptor (GR) and the pregnane X receptor (PXR) (Scheinman et al., 1995; Pei, 1996; Bertilsson et al., 1998;Karin, 1998; Kliewer et al., 1998; Lehmann et al., 1998). Ligand-bound GR interacts with either a positive GR or a negative GR response element, causing transactivation and trans-suppression of the target genes, respectively (Pei, 1996; Karin, 1998). Ligand-bound GR is also known to interact with other transcription factors and interfere with the transactivation by these proteins (Scheinman et al., 1995). Therefore, trans-suppression of ligand-bound GR can be achieved by either direct binding to a negative GR response element or inactivating other transcription factors. The PXR, a newly identified nuclear receptor, interacts with a cis-DNA response element and activates the transcription (Bertilsson et al., 1998; Kliewer et al., 1998; Lehmann et al., 1998). Although both receptors can be activated by dexamethasone, the concentration required for the activation is ∼100-fold different (Schustz et al., 1984;Schustz and Guzeliant, 1984; Jackson et al., 1998). The GR requires nanomolar, whereas the PXR requires micromolar, levels. In addition, only glucocorticoids activate the GR but both glucocorticoids and antiglucocorticoids activate the PXR (Bertilsson et al., 1998; Kliewer et al., 1998; Lehmann et al., 1998).
The suppression of rat hydrolase A, B, C, and S by dexamethasone is likely mediated by the GR, whereas the induction of HCE-1 and HCE-2 by this drug is mediated by the PXR. Several lines of evidence support this possibility. Suppression of the rat carboxylesterase gene expression requires only nanomolar levels of dexamethasone, whereas induction of human HCE-1 and HCE-2 requires micromolar levels (this study), suggesting that the suppression is mediated by a more sensitive pathway, namely, the GR involvement. Decreased activity towardpara-nitrophenylacetate and butanilicaine by several GR agonists is well correlated with the relative potency of these steroids in increasing the expression of the tyrosine aminotransferase (Schustz et al., 1984; Hosokawa et al., 1993; Jackson et al., 1998), a gene that is known to be regulated by the GR pathway. Suppressive effects are observed with dexamethasone but not pregnenolone 16α-carbonitrile (Hosokawa et al., 1993; Morgan et al., 1994a; Yan et al., 1995b), excluding the involvement of the PXR, which can be activated by both glucocorticoids and antiglucocorticoids. Both dexamethasone and rifampicin are shown to induce HCE-1 and HCE-2 with rifampicin being more efficacious; such a relative efficaciousness is observed in the PXR-mediated CYP3A4 induction (this study; Schustz et al., 1984; Schustz and Guzeliant, 1984), suggesting that the PXR not the GR is responsible for the induction of HCE-1 and HCE-2. The GR-mediated trans-suppression is achieved by either direct binding to a negative GR response element or interfering with other transcription factors (Scheinman et al., 1995; Pei, 1996; Karin, 1998). Recently, we have found that suppressive effects on the hydrolase A gene are seen only in the liver but not in the lung, an organ that is found to express high levels of this enzyme (D. Yang, L.M., and B.Y., unpublished results). These findings suggest that other liver nuclear proteins are required for the suppressive regulation and the interfering mechanism is involved in this process. However, it remains to be determined whether the difference in the disposition of this drug between these two organs contributes to such a tissue-differential regulation.
In summary, we report the regulation of carboxylesterase gene expression in cultured human and rat hepatocytes. Exposure of rat hepatocytes to dexamethasone results in a marked decrease of hydrolase A, B, and C. In contrast, exposure of human hepatocytes to this drug causes an increase in HCE-1 and HCE-2, two major forms of human liver microsomal carboxylesterases. The inductive effects in human hepatocytes are observed only when micromolar levels of dexamethasone are used. These results suggest that a major species difference exists regarding the regulation of carboxylesterase gene expression by dexamethasone. Alteration of carboxylesterase expression is clinically relevant, particularly in the activation of ester prodrugs. Camptothecin derivatives, a new class of topoisomerase I inhibitors, are increasingly used for several types of advanced cancers (Stucky-Marshall, 1999). However, only the corresponding hydrolytic metabolites are pharmacologically active. Carboxylesterase inhibitors are shown to significantly decrease the conversion, and decreased hydrolytic activity in patients is implicated to the decreased effectiveness of these drugs in chemotherapy (Ogasawara et al., 1995;Ewesuedo and Ratain, 1997; Saltz, 1997; Stucky-Marshall, 1999).
Footnotes
-
Send reprint requests to: Dr. Bingfang Yan, Department of Biomedical Sciences, University of Rhode Island, Kingston, RI 02881. E-mail: byan{at}uri.edu
-
↵1 Equal contributions were made by these authors.
-
↵2 Present address: Department of Drug Delivery and Disposition, School of Pharmacy, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599.
-
This work was partially supported by National Institute of Environmental Health Sciences (NIEHS) Grant ES07965 and a New Investigator Award from the American Association of Colleges of Pharmacy.
- Abbreviations used are::
- CYP
- cytochrome P-450
- DMEM
- Dulbecco's modified Eagle's medium
- GR
- glucocorticoid receptor
- PCR
- polymerase chain reaction
- PXR
- pregnane X receptor
- PAGE
- polyacrylamide gel electrophoresis
- TNM-FH
- Trichoplusia ni Medium-Formulation Hink
- Received July 12, 1999.
- Accepted October 13, 1999.
- The American Society for Pharmacology and Experimental Therapeutics