Abstract
Human arylacetamide deacetylase (AADAC) catalyzes the hydrolysis of some clinically used drugs, but the information available on its substrates is limited. To increase our knowledge of the AADAC substrates, we examined whether AADAC catalyzes the hydrolysis of indiplon, which was initially developed as a hypnotic sedative drug. It has been reported that approximately 30–40% of the administered indiplon was hydrolyzed to deacetylindiplon in humans, but the enzyme responsible for this hydrolysis had not been identified. We detected high levels of indiplon hydrolase activity in human liver microsomes (HLMs), but the levels found in human liver cytosol and plasma were scarcely detectable. Recombinant AADAC showed a high level of indiplon hydrolase activity, whereas recombinant carboxylesterase 1 (CES1) and 2 (CES2) showed marginal activity. The indiplon hydrolase activity of HLM was potently inhibited by vinblastine, a potent inhibitor of AADAC and CES2, but it was not inhibited by digitonin and telmisartan, inhibitors of CES1 and CES2, respectively. In a panel of 24 individual HLM samples, the indiplon hydrolase activities were significantly correlated with the hydrolase activities of flutamide, phenacetin, and rifampicin, which are known AADAC substrates. An HLM sample with a homozygous AADAC*3 allele, which was previously found to exhibit decreased enzyme activity, showed the lowest indiplon hydrolase activity among the 24 tested samples. Collectively, we found that human AADAC is responsible for the hydrolysis of indiplon. Thus, we can add indiplon to the list of human AADAC substrates.
Introduction
Drug-metabolizing enzymes play roles in the detoxification of drugs and the activation of prodrugs (Meyer, 1996; Begg et al., 2012). Esterases contribute to the hydrolysis of 10% of the clinical therapeutic drugs that contain ester, amide, and thioester bonds. Among the esterases, carboxylesterases (CESs) are the best studied and recognized enzymes that catalyze the hydrolysis of various xenobiotic and endogenous compounds (Fukami and Yokoi, 2012). In humans, CES1 and CES2 isoforms mainly contribute to drug hydrolysis and exhibit different tissue distributions and substrate specificities. CES1 is expressed predominantly in the human liver, whereas CES2 is expressed in the liver and in extrahepatic tissues, including the gastrointestinal tract and kidney (Xu et al., 2002; Imai et al., 2006). CES1 prefers substrates with a small alcohol moiety and a large acyl moiety, whereas CES2 prefers those with a large alcohol moiety and small acyl moiety (Imai et al., 2006).
Arylacetamide deacetylase (AADAC) was first identified as an enzyme that catalyzes the hydrolysis of 2-acetylaminofluorene (Probst et al., 1991). AADAC is expressed mainly in the human liver and gastrointestinal tract (Watanabe et al., 2009) and is localized on the endoplasmic reticulum membrane (Frick et al., 2004). We recently discovered that AADAC is responsible for the hydrolysis of drugs, such as flutamide (Watanabe et al., 2009), phenacetin (Watanabe et al., 2010), and rifamycins (rifampicin, rifabutin, and rifapentine) (Nakajima et al., 2011). The common structural characteristics of these identified substrates of human AADAC include the existence of a small acyl moiety, such as an acetyl or isopropyl group. However, the types of drugs that can be substrates of human AADAC are largely unknown. In this study, to increase the available information on AADAC substrates, we investigated whether AADAC can hydrolyze indiplon, which possesses an acetyl group.
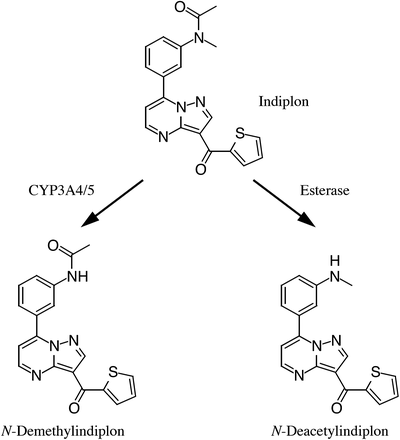
Indiplon, which shows a higher affinity and selectivity to GABAA receptors compared with zolpidem and zaleplon, was initially developed as a nonbenzodiazepine sedative-hypnotic agent (Petroski et al., 2006). Indiplon was expected to be a safer and more profitable drug than the currently prescribed drugs, but its development was discontinued owing to the company’s circumstances. The metabolic pathways of indiplon were investigated during the early stages of its development. Indiplon is mainly metabolized to N-demethylindiplon by CYP3A4/5 and is also hydrolyzed to N-deacetylindiplon in human liver microsomes (HLMs) (Fig. 1) (Madan et al., 2007). The N demethylation reaction accounts for 60–70% of the overall in vitro clearance of indiplon in HLMs, whereas the N-deacetylation (hydrolysis) process accounts for 30–40% (Madan et al., 2007). Indiplon hydrolysis occurs in HLM and is inhibited by bis(p-nitrophenyl) phosphate (BNPP), a general inhibitor of serine esterases, but not by phenylmethylsulfonyl fluoride (PMSF) (Madan et al., 2007), a potent inhibitor of CES enzymes (Xie et al., 2002). Indiplon possesses an acetyl group in its structure. This background information prompted us to investigate whether human AADAC can catalyze indiplon hydrolysis.
Metabolic pathways of indiplon in humans.
Materials and Methods
Chemicals and Reagents.
Digitonin, diisopropylphosphorofluoride (DFP), flutamide, loperamide, p-nitrophenol, 4-nitro-3-(trifluoromethyl)phenylamine, phenacetin, PMSF, physostigmine sulfate (eserine), rifampicin, and vinblastine were purchased from Wako Pure Chemical Industries (Osaka, Japan). BNPP, fenofibrate, p-nitrophenyl acetate (PNPA), and p-phenetidine were purchased from Sigma-Aldrich (St. Louis, MO). Indiplon was purchased from Carbosynthe (Berkshire, UK). Telmisartan was purchased from LKT Laboratories (St. Paul, MN). 25-Deacetylrifampicin, fenofibric acid, irinotecan hydrochloride, and 7-ethyl-10-hydroxycamptotesin were purchased from Toronto Research Chemicals (Toronto, Canada). Recombinant human AADAC, CES1, and CES2 expressed in baculovirus-infected insect cells were prepared as previously described (Fukami et al., 2010; Watanabe et al., 2010). The homogenates of COS7 cells, fibroblast-like cell line from monkey kidney tissue, expressing AADAC.1, AADAC.2, and AADAC.3 were prepared as in our previous study (Shimizu et al., 2012). All other chemicals used in this study were of analytical or the highest quality commercially available.
Human Liver and Plasma.
Pooled HLMs (n = 50) and pooled human liver cytosol (n = 50) were purchased from Corning (Corning, NY). Individual HLMs from 24 donors (12 men, 12 women) were supplied by the National Disease Research Interchange (Philadelphia, PA) through the Human and Animal Bridging Research Organization (Chiba, Japan). Blood was collected from 50 healthy volunteers who had been taking no medications (age 21–53 years; men, n = 31; women, n = 19). The blood, combined with heparin as an anticoagulant, was allowed to stand for 30 minutes and was then centrifuged at 2000g for 15 minutes to separate the plasma. The use of human tissue and plasma samples was approved by the ethics committees of Kanazawa University (Kanazawa, Japan).
Indiplon Hydrolase Activity.
The indiplon hydrolase activity was determined as follows: a typical incubation mixture (final volume, 0.2 ml) contained 100 mM potassium phosphate buffer (pH 7.4) and various enzyme sources (HLMs, human liver cytosol, and homogenates of Sf21 cells expressing AADAC, CES1, or CES2: 0.4 mg/ml; human plasma: 2.5 µl). Indiplon was dissolved in DMSO to obtain a final concentration of dimethyl sulfoxide (DMSO) in the incubation mixture of 1.0%. The reaction was initiated by the addition of indiplon (final concentration, 50 µM) after a 2-minute preincubation at 37°C. After a 30-minute incubation at 37°C, the reaction was terminated by the addition of 100 µl of ice-cold acetonitrile. It was confirmed that the rate of deacetylindiplon formation was linear up to a protein concentration of 1.0 mg/ml and up to an incubation time of 60 minutes. After removal of the protein by centrifugation at 10,000 rpm for 5 minutes, a 50-μl aliquot of the supernatant was subjected to high-performance liquid chromatography (HPLC). The HPLC equipment consisted of an L-7100 pump (Hitachi, Tokyo, Japan), an L-7200 autosampler (Hitachi), an L-7405 UV detector (Hitachi), and a D-2500 HPLC Chromato-Integrator (Hitachi) equipped with a Symmetry Shield RP8 (4.6 × 150 mm ID, 5 μm; Waters, Tokyo, Japan). The eluent was monitored at 230 nm with a noise-base clean Uni-3 (Union, Gunma, Japan). The mobile phase was 23% acetonitrile containing 0.1% trifluoroacetic acid, and the flow rate was 1.0 ml/min. The column temperature was 35°C.
Identification and Quantification of Deacetylindiplon.
We used liquid chromatography–tandem mass spectometry (LC-MS/MS) analysis to determine whether the identified peak represents deacetylindiplon because the authentic standard was not commercially available. The LC equipment comprised an HP1100 system with a binary pump, an automatic sampler, and a column oven (Agilent Technologies, Santa Clara, CA) equipped with Inertsil ODS-3 (2.1 × 50 mm, ID 5 μm; GL Science, Tokyo, Japan). The column temperature was 25°C. The mobile phase was 0.1% trifluoroacetic acid (A) and acetonitrile with 0.1% trifluoroacetic acid (B). The conditions for elusion were as follows: 20% B (0–10 minutes), 20–25% B (10–11 minutes), and 25% B (11–30 minutes). The flow rate was 0.2 ml/min. The LC was connected to a PE Sciex API2000 tandem mass spectrometer (AB Sciex, Framingham, MA), which was operated in the positive electrospray ionization mode. In the single-quadrupole mode (Q1-scan), full-scan spectra were acquired with a scan range of m/z 80–400. In the multiple-reaction monitoring (MRM) mode, 2 m/z ion transitions (m/z 377.1 and 293.2 for indiplon and m/z 335.1 and 251.2 for deacetylindiplon) were monitored. The turbo gas was maintained at 550°C. Nitrogen was used as the nebulizing, turbo, and curtain gas at 70, 40, and 50 psi, respectively. The parent or fragment ions were filtered in the first quadrupole and dissociated in the collision cell using nitrogen as the collision gas. The collision energy was 51 V for indiplon. The analytical data were processed using the Analyst software (version 1.5; Applied Biosystems, Foster City, CA).
The deacetylindiplon formed was quantified by HPLC analysis based on the decreased amount of indiplon. In detail, indiplon at concentrations of 0.5, 1, 1.5, and 2 µM was incubated with HLMs (1.0 mg/ml). The decreased amounts of indiplon were quantified and applied to the peaks of deacetylindiplon formed in the reaction. After determining the peak height per known content of deacetylindiplon, the value was used to calculate the amount of deacetylindiplon formed in the incubation mixture.
Kinetic Analysis of the Indiplon Hydrolase Activity of HLMs and Recombinant AADAC.
A kinetic analysis was performed using HLMs and recombinant AADAC with substrate concentrations ranging from 20 to 500 µM. The kinetic parameters were estimated from the curve fitted to a Michaelis-Menten equation using a computer program designed for nonlinear regression analysis (KaleidaGraph; Synergy Software, Reading, PA).
Inhibition Analysis of Indiplon Hydrolase Activity.
To examine the involvement of AADAC in indiplon hydrolysis, an inhibition analysis was performed. BNPP, DFP, and PMSF are serine esterase inhibitors. Eserine, a cholinesterase inhibitor, inhibits AADAC and CES2 (Kobayashi et al., 2012a). We recently found that vinblastine inhibits AADAC and CES2 and that digitonin and telmisartan inhibit CES1 and CES2, respectively (M. Shimizu, T. Fukami, M. Nakajima, and T. Yokoi, unpublished data). Loperamide specifically inhibits CES2 (Quinney et al., 2005). The concentrations of the inhibitors were set to the appropriate conditions showing inhibition specificities as follows: BNPP, DFP, PMSF, and eserine, 100 µM; vinblastine, 50 µM; digitonin, 200 µM; telmisartan and loperamide, 5 µM. PMSF, vinblastine, and digitonin were dissolved in DMSO, and telmisartan was dissolved in methanol such that the final concentration of the organic solvent in the incubation mixture was 1.5%. The other inhibitors were dissolved in distilled water. In the presence of these inhibitors, the activity of indiplon hydrolase was measured as described already herein.
Effects of AADAC Genetic Polymorphisms on Indiplon Hydrolase Activity.
We previously found polymorphic alleles of the human AADAC gene (Shimizu et al., 2012): AADAC*2 and AADAC*3. The AADAC proteins encoded by AADAC*2 and AADAC*3 are AADAC.2 and AADAC.3. The AADAC protein from wild-type AADAC*1 is AADAC.1. Recombinant AADAC.2 and AADAC.3, prepared as in our previous study (Shimizu et al., 2012), were used to determine the effects of genetic polymorphisms of the AADAC gene on the indiplon hydrolase activity. In addition, the indiplon hydrolase activity of the individual HLM samples, the AADAC genotype of which was previously determined (Shimizu et al., 2012), was also measured.
Other Hydrolase Activities.
The PNPA, flutamide, phenacetin, and rifampicin hydrolase activities were determined according to our previous reports (Watanabe et al., 2009, 2010; Nakajima et al., 2011; Shimizu et al., 2012). Fenofibrate and irinotecan hydrolase activities were also determined according to our previous report (Takahashi et al., 2009; Fukami et al., 2010). Individual HLMs from 24 donors were used as the enzyme sources. The protein concentrations were as follows: PNPA hydrolase activity, 0.1 mg/ml; fenofibrate and irinotecan hydrolase activities, 0.2 mg/ml; flutamide and phenacetin hydrolase activities, 0.4 mg/ml; rifampicin hydrolase activity, 0.5 mg/ml. The concentrations of PNPA, fenofibrate, irinotecan, flutamide, phenacetin, and rifampicin were 500 µM, 10 µM, 2 µM, 500 µM, 1 mM, and 50 µM, respectively.
Statistical Analysis.
The statistical significance between multiple groups was determined by analysis of variance followed by Dunnett’s test using the Instat 2 software (GraphPad Software, San Diego, CA). The correlation analyses were performed using the Spearman rank method. A value of P < 0.05 was considered statistically significant.
Results
Indiplon Hydrolase Activities of HLMs, Human Liver Cytosol and Plasma, and Recombinant Enzymes.
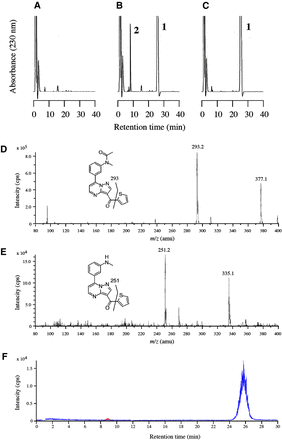
In the HPLC analysis, the retention time of indiplon was 25 minutes. When indiplon was incubated with HLMs, a single peak at the retention time of 8.3 minutes appeared (Fig. 2B), and this peak was not affected by the other components of HLMs (Fig. 2A). This peak disappeared when a general esterase inhibitor DFP (100 µM) was added to the reaction mixture (Fig. 2C). We surmised that the peak at 8.3 minutes corresponds to deacetylindiplon. To confirm this assumption, LC-MS/MS was performed. Through Q1-scan monitoring at m/z 80–400, two peaks at almost the same retention times as those observed in the HPLC analysis (approximately 8.5 and 26 minutes) were observed. The mass spectrum of the peak at 26 minutes showed ions m/z 377.1, which corresponds to indiplon, and m/z 293.2, which most likely corresponds to the product derived through the loss of the thiophene group (Fig. 2D). The mass spectrum of the peak at 8.5 minutes showed ions m/z 335.1, which corresponds to deacetylindiplon, and m/z 251.2, which most likely corresponds to the product after the loss of the thiophene group (Fig. 2E). Fig. 2F shows the MRM chromatogram monitored at m/z 337.1/293.2 and 335.1/251.2. We also confirmed that the peak in MRM at m/z 335.1/251.2 disappeared when DFP was added to the reaction mixture (data not shown). These results suggest that the peak at 8.3 minutes is a hydrolyzed metabolite of indiplon, namely, deacetylindiplon.
Representative HPLC chromatograms (A–C) and LC-MS/MS spectra (D and E) and chromatogram (F) of indiplon and deacetylindiplon. HLMs were incubated with (A) no indiplon, (B, D, E, and F) 50 µM indiplon, and (C) 50 µM indiplon plus 100 µM DFP, an inhibitor of serine esterase. (A–C) Peak 1, indiplon; peak 2, deacetylindiplon. The Q1 scan with a scan range of m/z 80–400 showed two peaks at (D) approximately 26 minutes (spectra showed ions m/z 377.1 and 293.2) corresponding to indiplon and (E) 8.5 minutes (spectra showed ions m/z 335.1 and 251.2) corresponding to deacetylindiplon. The MRM spectra at (F) m/z 337.1 and 293.2 (corresponding to indiplon, blue) and m/z 335.1 and 251.2 (corresponding to deacetylindiplon, red) were obtained.
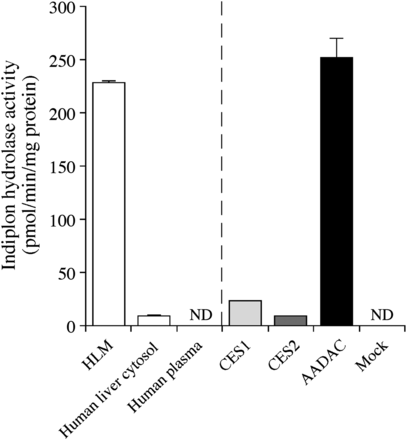
The indiplon hydrolase activity of HLMs was 228.0 ± 2.2 pmol/min per milligram of protein at a substrate concentration of 50 µM (Fig. 3). The activity of human liver cytosol (9.4 ± 0.8 pmol/min per milligram of protein) was markedly lower than that of HLMs. Human plasma showed no activity, indicating that the enzymes localized in plasma, such as butyrylcholinesterase (BCHE) and paraoxonase (PON), are not involved in indiplon hydrolysis. The AADAC and CES enzymes, which are expressed in the endoplasmic reticulum, are considered to catalyze indiplon hydrolysis, although the CES enzymes are moderately expressed in the cytosolic fraction. Interestingly, recombinant AADAC showed substantial indiplon hydrolase activity, whereas recombinant CES1 and CES2 showed marginal activity. These results suggest that indiplon is mainly hydrolyzed by AADAC in HLM.
Indiplon hydrolase activities of HLMs, human liver cytosol and plasma, and recombinant esterases. The substrate concentration was 50 µM. Each column represents the mean ± S.D. of triplicate determinations. ND, Not detected.
Kinetic Analyses of the Indiplon Hydrolase Activities of HLMs and Recombinant AADAC.
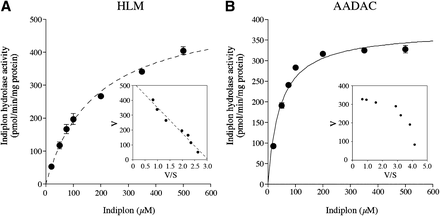
A kinetic analysis was performed using HLMs (Fig. 4A) and recombinant AADAC (Fig. 4B). Both were fitted to Michaelis–Menten kinetics, and the calculated kinetic parameters are shown in Table 1. The Km value found for recombinant AADAC was slightly lower than that found for HLMs, and this difference may be due to the difference in membrane environment in liver and Sf21 cells.
Kinetic analyses of indiplon hydrolase activities of (A) HLMs and (B) recombinant AADAC. An Eadie–Hofstee plot is shown in the inset. The kinetic parameters were estimated from the fitted curve using the KaleidaGraph computer program designed for nonlinear regression analysis. Each data point represents the mean ± S.D. of triplicate determinations.
Kinetic parameters of indiplon hydrolase activities
Inhibitory Patterns of the Indiplon Hydrolase Activity of HLM and Recombinant AADAC.
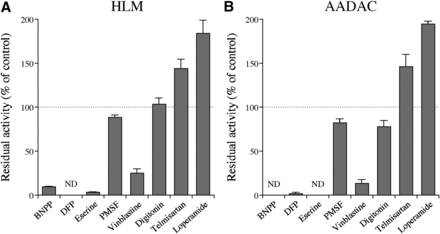
An inhibition study was performed to examine whether AADAC is the enzyme responsible for indiplon hydrolysis in the human liver (Fig. 5). The indiplon hydrolase activity of HLMs was inhibited by BNPP, DFP, and eserine but not by PMSF. We previously reported that the inhibitory effects of PMSF on AADAC were weak (Watanabe et al., 2009) and that eserine, a potent inhibitor of cholinesterase, potently inhibits AADAC and CES2 (Kobayashi et al., 2012a). In addition, the indiplon hydrolase activity of HLM was potently inhibited by vinblastine, an inhibitor of AADAC and CES2 (Fig. 5A) but not by digitonin (CES1 inhibitor). It was hypothesized that the inhibition by vinblastine was due to the inhibition of AADAC. Unexpectedly, the activity of HLM was activated by telmisartan and loperamide. Of particular note was that the inhibition or activation pattern found for HLM is quite similar to that found for recombinant AADAC (Fig. 5B). The results suggest that AADAC is primarily responsible for indiplon hydrolysis in the human liver.
Effects of chemical inhibitors on indiplon hydrolase activities of (A) HLMs and (B) recombinant AADAC. The substrate concentration was 50 µM. The concentrations of the inhibitors were as follows: BNPP, DFP, PMSF, and eserine, 100 µM; vinblastine, 50 µM; digitonin, 200 µM; telmisartan and loperamide, 5 µM. The control activities were (A) 149.2 and (B) 71.6 pmol/min per milligram of protein, respectively. Each column represents the mean ± S.D. of triplicate determinations. ND, not detected.
Effects of AADAC Genetic Polymorphisms on Indiplon Hydrolase Activity.
The AADAC*2 allele has a single nucleotide polymorphism of c.841G>A, which leads to an amino acid substitution of V281I, and the AADAC*3 allele has two single nucleotide polymorphisms of c.841G>A and c.1288T>C, leading to the amino acid changes of V281I and ×400Q, resulting in the extension of the C terminal. We previously reported that the enzyme activity of AADAC.2 is almost the same as that of AADAC.1; however, the enzyme activity of AADAC.3 is prominently lower than that of AADAC.1 (Shimizu et al., 2012). In this study, we evaluated the indiplon hydrolase activities of recombinant AADAC variants (Fig. 6A). The activities were normalized with the relative expression level of AADAC protein (AADAC.1: 1.0 U/mg of protein, AADAC.2: 0.9 ± 0.0 U/mg of protein, AADAC.3: 0.5 ± 0.0 U/mg protein, respectively), which was previously determined through immunoblot analysis (Shimizu et al., 2012). Compared with AADAC.1 (91.7 ± 5.7 pmol/min per unit), AADAC.2 showed slightly lower activity (64.7 ±1.8 pmol/min per unit) and AADAC.3 showed markedly lower activity (7.7 ± 1.8 pmol/min per unit).
(A) Indiplon hydrolase activities of homogenates of COS7 cells expressing human wild-type and variant AADAC. The substrate concentration was 50 μM. The activities were normalized with the AADAC protein level through immunoblot analysis (AADAC.1: AADAC.2: AADAC.3 = 1.0 ± 0.0 U/mg of protein: 0.9 ± 0.0 U/mg of protein: 0.5 ± 0.0 U/mg of protein). Each column represents the mean ± S.D. of triplicate determinations. *P < 0.05 compared with AADAC.1. ND, not detected. (B) Indiplon hydrolase activities of 24 individual HLMs with different AADAC genotypes. The substrate concentration was 50 μM.
We then measured the indiplon hydrolase activities in microsomes from 24 individual livers, the AADAC gene of which had been previously genotyped (Shimizu et al., 2012) (Fig. 6B). The indiplon hydrolase activities varied from 16.8 to 143.4 pmol/min per milligram of protein, showing 8.5-fold variation. The numbers of samples genotyped as AADAC*1/AADAC*1, AADAC*1/AADAC*2, AADAC*2/AADAC*2, and AADAC*3/AADAC*3 were 1, 13, 9, and 1, respectively. The indiplon hydrolase activity of HLM from a subject genotyped as AADAC*3/AADAC*3 (16.8 pmol/min per milligram of protein) was the lowest among all the HLM samples used in this study (AADAC*1/AADAC*1: 28.9 pmol/min per milligram of protein, AADAC*1/AADAC*2: 76.4 ± 34.0 pmol/min per milligram of protein, AADAC*2/AADAC*2: 65.9 ± 36.6 pmol/min per milligram of protein). The limitations of this study were that only one homozygote of AADAC*1 was tested and that the interindividual variability within the same AADAC genotype group was large; however, it appears that the AADAC genetic polymorphisms may be a cause of the interindividual variability observed in the indiplon hydrolase activity.
Correlation Analyses between the Hydrolase Activities of Indiplon and Known AADAC Substrates.
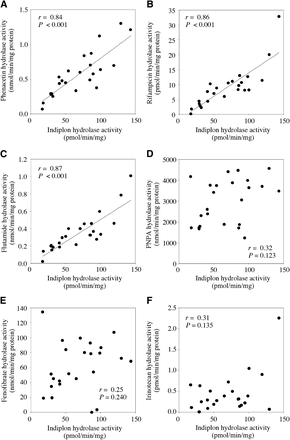
To verify the contribution of AADAC to indiplon hydrolysis, we performed correlation analyses (Fig. 7). Phenacetin and rifampicin are specific substrates for human AADAC (Watanabe et al., 2010; Nakajima et al., 2011). Although CES2 also contributes to flutamide hydrolysis, we previously found that its activity at 500 µM reflects the AADAC enzyme activity (Kobayashi et al., 2012b). PNPA is hydrolyzed by multiple esterases, including the CES enzymes (Watanabe et al., 2009). Fenofibrate and irinotecan are selected as marker probes for CES1 and CES2, respectively (Fukami and Yokoi, 2012; Kobayashi et al., 2012b). The indiplon hydrolase activity was significantly correlated (P < 0.001) to the phenacetin (r = 0.84), rifampicin (r = 0.86), and flutamide hydrolase activities (r = 0.87). In contrast, no correlation was observed with the hydrolase activities of PNPA (r = 0.32, P = 0.123), fenofibrate (r = 0.25, P = 0.240), and irinotecan (r = 0.31, P = 0.135). Collectively, AADAC was demonstrated to be the enzyme responsible for indiplon hydrolysis.
Correlation analyses between indiplon and (A) phenacetin, (B) rifampicin, (C) flutamide, (D) PNPA, (E) fenofibrate, and (F) irinotecan hydrolase activities of individual HLM samples from 24 donors. These analyses were performed using the Spearman rank correlation method.
Discussion
Esterases play important roles in the detoxification and pharmacological activation of clinical drugs. It is well known that the CES enzymes, namely CES1 and CES2, catalyze the hydrolysis of a variety of drugs. We recently found that AADAC is also involved in the hydrolysis of several drugs (Watanabe et al., 2009, 2010; Nakajima et al., 2011). However, its substrate specificity remains unclear because of its limited number of substrates. Indiplon, which had been initially developed as a hypnotic sedative drug, is hydrolyzed to N-deacetylindiplon (Madan et al., 2007), but the enzyme responsible for this hydrolysis remained unknown. In this study, we investigated whether human AADAC catalyzes indiplon hydrolysis.
The indiplon hydrolase activity was measured using human liver preparations (HLMs and liver cytosol) and plasma (Fig. 3). HLMs showed substantial activity, and human liver cytosol and plasma showed scarce activity (Fig. 3). AADAC is localized in the endoplasmic reticulum (Frick et al., 2004), whereas the CES1 and CES2 enzymes exist in both the endoplasmic reticulum and the cytosol (Xu et al., 2002; Tabata et al., 2004). The CES enzymes are not present in human plasma (Li et al., 2005). There is no direct evidence for the presence of the AADAC protein in human plasma, and our previous study demonstrated that phenacetin hydrolysis catalyzed by AADAC is not detected in human plasma (Watanabe et al., 2010). These results suggest that indiplon is hydrolyzed by AADAC, and this hypothesis was confirmed using recombinant enzymes (Fig. 3). BCHE and PON are other esterases involved in drug metabolism. These esterases are expressed in the human liver (Jbilo et al., 1994; Reddy et al., 2001) and plasma (Chatonnet and Lockridge, 1989; Reddy et al., 2001). Because indiplon hydrolysis was not observed in human plasma (Fig. 3), the involvement of BCHE and PON was excluded.
To confirm more clearly the role of AADAC in indiplon hydrolysis, an inhibition analysis was performed (Fig. 5). The indiplon hydrolase activity of HLM was potently inhibited by vinblastine and eserine (Fig. 5A), which were previously found to inhibit AADAC and CES2 (M. Shimizu, T. Fukami, M. Nakajima, and T. Yokoi, unpublished data; Kobayashi et al., 2012a). However, indiplon hydrolysis was not inhibited by telmisartan and loperamide, which inhibit CES2 (Shimizu et al., submitted; Quinney et al., 2005). Thus, the inhibition of indiplon hydrolysis by vinblastine and eserine is likely the result of the inhibition of AADAC activity. Moreover, the activity was inhibited by BNPP and DFP but not by PMSF. Madan et al. (2007) also reported that the indiplon hydrolysis of HLMs is potently inhibited by BNPP but not by PMSF, which is in agreement with our results. Together with the similarity in the inhibitory characteristics between HLM and recombinant AADAC (Fig. 5B), the results strengthened the conclusion that AADAC is the enzyme responsible for indiplon hydrolysis in the human liver. The indiplon hydrolase activity of HLMs and recombinant AADAC was activated by telmisartan and loperamide (Fig. 5). Such heterotropic effects were previously demonstrated for flutamide, which enhances the phenacetin hydrolase activity of AADAC (Watanabe et al., 2010). Therefore, AADAC may have a large binding site or multiple binding sites for compounds that result in an enhancement in its activity. However, loperamide does not activate AADAC-catalyzed phenacetin hydrolysis, and flutamide does not activate AADAC-catalyzed rifampicin hydrolysis (A. Watanabe and T. Fukami, unpublished data). Thus, the heterotropic effects of AADAC appear to be dependent on the combination of substrates and effectors.
There are genetic polymorphisms in the human AADAC gene. For indiplon hydrolysis, we found that AADAC.2 showed slightly lower activity and that AADAC.3 showed remarkably lower activity than AADAC.1 (Fig. 6A). This tendency is consistent with the results obtained for flutamide, phenacetin, and rifampicin hydrolysis in our previous study (Shimizu et al., 2012). In addition, HLMs from a subject genotyped as AADAC*3/AADAC*3 showed the lowest indiplon hydrolase activity among the 24 HLM samples tested (Fig. 6B). However, the limited sample size for each genotype precluded a statistical analysis. We previously found that AADAC protein expression levels in these HLM samples were correlated with the AADAC enzyme activities measured by using flutamide, phenacetin, and rifampicin as substrates (Shimizu et al., 2012). An HLM sample from a subject genotyped with AADAC*1/AADAC*1 showed low AADAC expression (Shimizu et al., 2012); therefore, its indiplon hydrolase activity would be relatively low. A possible reason might be that other genetic mutations exist in this sample to decrease the AADAC expression level. Further study with an increased number of samples will provide insights into the effects of AADAC genetic polymorphisms on indiplon hydrolase activity. In the panel of 24 individual HLM samples, the indiplon hydrolase activities were significantly correlated with the hydrolase activities of phenacetin, rifampicin, and flutamide, which are substrates of AADAC (Fig. 7). These results support the conclusion that indiplon hydrolysis is catalyzed by AADAC.
The present findings support our previous conjecture that AADAC prefers compounds with a small acyl moiety, such as an acetyl or isobutyryl group, as substrates (Watanabe et al., 2009, 2010; Nakajima et al., 2011). It is recognized that CES1 prefers substrates with a small alcohol moiety and large acyl moiety, whereas CES2 prefers substrates with a large alcohol moiety and small acyl moiety (Imai et al., 2006). AADAC may show a substrate specificity similar to that of CES2. The expansion of the number of AADAC substrates is a subject of future study and will help us understand the pharmacological and toxicological significance of AADAC in pharmacotherapy.
In conclusion, we found that indiplon is hydrolyzed by AADAC in humans. This study may expand our knowledge on the substrate preference of human AADAC.
Authorship Contributions
Participated in research design: Shimizu, Fukami, Nakajima.
Conducted experiments: Shimizu, Fukami, Ito, Kurokawa, Kariya.
Contributed new reagents or analytic tools: Shimizu.
Performed data analysis: Shimizu, Fukami.
Wrote or contributed to the writing of the manuscript: Shimizu, Fukami, Nakajima, Yokoi.
Footnotes
- Received November 24, 2013.
- Accepted January 23, 2014.
This work was supported in part by a Grant-in-Aid for Scientific Research (C) from the Japan Society for the Promotion of Science [23590174].
Abbreviations
- AADAC
- arylacetamide deacetylase
- BCHE
- butyrylcholinesterase
- BNPP
- bis(p-nitrophenyl) phosphate
- CES
- carboxylesterase
- DFP
- diisopropylphosphorofluoride
- DMSO
- dimethyl sulfoxide
- HLM
- human liver microsome
- HPLC
- high-performance liquid chromatography
- LC-MS/MS
- liquid chromatography–tandem mass spectrometry
- MRM
- multiple-reaction monitoring
- PMSF
- phenylmethylsulfonyl fluoride
- PNPA
- p-nitrophenyl acetate
- PON
- paraoxonase
- Copyright © 2014 by The American Society for Pharmacology and Experimental Therapeutics