Abstract
Polymorphisms in UGT1A9 were associated with reduced toxicity and increased response to irinotecan in cancer patients. UDP-glucuronosyltransferase (UGT) protein expression, glucuronidation activities for 7-ethyl-10-hydroxycamptothecin (SN-38), and probe substrates of the UGT1A9 and UGT1A1 were measured in 48 human livers to clarify the role of UGT1A9 variants on the in vitro glucuronidation of SN-38. Genotypes were assessed for UGT1A9 (–2152C>T, –275T>A, and –118T9>10), three novel UGT1A9 variants (–5366G>T, –4549T>C, and I399C>T), and UGT1A1 (–53TA6>7, –3156G>A, and –3279T>G). Of all the variants, the UGT1A9 I399C>T was associated with the most dramatic change in SN-38-glucuronide (SN-38G) (2.64-fold; p = 0.0007). Compared with UGT1A9 I399C/C, homozygous I399T/T presented elevated UGT1A1 and UGT1A9 proteins and higher glucuronidation of UGT1A9 and UGT1A1 substrates (p < 0.05). The very low linkage disequilibrium (r2 < 0.19) between UGT1A9 I399 and all the other UGT1A1 and UGT1A9 variants suggests a direct effect or linkage to unknown functional variant(s) relevant to SN-38 glucuronidation. The UGT1A9 –118T9/10 was also linked to alteration of SN-38 glucuronidation profiles in the liver (p < 0.05) and was associated with higher UGT1A1 protein (p = 0.03). However, UGT1A9 –118T10 appears to have low functional impact as a result of the lack of correlation with UGT1A9 protein levels and a modest 1.4-fold higher reporter gene expression associated with the –118T10 allele in HepG2 cells (p = 0.004). In contrast, the UGT1A9 –5366T, –4549C, –2152T, and –275A, associated with higher UGT1A9 protein (2-fold; p < 0.05), have no influence on SN-38G. Despite limitations resulting from sample size, results indicate that UGT1A9 I399 and –118T9/10 may represent additional candidates in combination with UGT1A1 promoter haplotypes for the prediction of SN-38 glucuronidation profile in vivo.
Irinotecan (or CPT-11) is a topoisomerase inhibitor used to treat metastatic colorectal cancer (Douillard et al., 2000; Saltz et al., 2000). Its metabolism is complex and includes oxidation by cytochrome P450 and activation by carboxylesterase to form the active metabolite SN-38 (Kawato et al., 1991). Glucuronidation catalyzed by UDP-glucuronosyltransferases (UGTs) represents the major inactivation pathway for SN-38 (Gupta et al., 1994). Of the 16 functional human UGT enzymes, UGT1A1 and UGT1A9 were identified as the main hepatic enzymes involved in the inactivation of SN-38 to SN-38-glucuronide (SN-38G) (Hanioka et al., 2001; Gagne et al., 2002). UGT1A1 and UGT1A9 are encoded by a single gene by exon sharing of individual exons 1 with common exons 2 to 5, whereas at least six other UGT1A proteins originate from this gene (Gong et al., 2001).
Several studies revealed the importance of UGT1A1 and UGT1A9 in the hepatic metabolism of SN-38 (Iyer et al., 1998; Gagne et al., 2002; Carlini et al., 2005). A number of polymorphisms in UGT1A1 and UGT1A9 affecting expression and protein function have been identified and could potentially modulate the metabolism of SN-38 in vivo (Zheng et al., 2001; Guillemette, 2003; Villeneuve et al., 2003; Girard et al., 2004; Yamanaka et al., 2004), as well as response to irinotecan-based chemotherapy (Ando et al., 2000; Innocenti et al., 2004; Marcuello et al., 2004; Rouits et al., 2004; Carlini et al., 2005).
Among these genetic variations, patients with the UGT1A1*28 variant, associated with Gilbert's syndrome (Bosma et al., 1995), show higher levels of SN-38 and experienced severe diarrhea and neutropenia (Ando et al., 2000; Innocenti et al., 2004; Marcuello et al., 2004; Rouits et al., 2004). Additional polymorphisms in the UGT1A1 gene have been reported and linked to a variable SN-38 glucuronidation, namely, the –3156G>A and the –3279T>G variations (Innocenti et al., 2002; Sugatani et al., 2002). Genotyping of these variations in the promoter region of UGT1A1, along with the –53 variant, has been suggested to improve the prediction of UGT1A1 status, and specific UGT1A1 haplotypes are associated with altered SN-38 phenotypes (Innocenti et al., 2002, 2005; Sai et al., 2004; Kitagawa et al., 2005).
Because the UGT1A9 protein is involved in the formation of SN-38G (Hanioka et al., 2001; Gagne et al., 2002), we recently identified polymorphic variations in the Caucasian population associated with altered gene expression or catalytic efficiency, namely, the –2152C>T, –275T>A, and M33T(98T>C) (Villeneuve et al., 2003; Girard et al., 2004). Another group reported that the –118T9>10 repeat was associated with higher reporter gene expression in HepG2 liver cells (Yamanaka et al., 2004), and this variant was recently linked to higher incidence toxicity and increased response to irinotecan in cancer patients. (Carlini et al., 2005). Still, definitive conclusions on the exact role of UGT1A1 and UGT1A9 variations and their influence on irinotecan response in patients cannot be drawn. Additional studies are critical for determining the role of other polymorphisms in UGTs, in addition to UGT1A1, and especially the importance of UGT1 haplotypes.
In this study, we wanted to gain insights into the role of polymorphisms in the UGT1A9 in modulating SN-38G formation in the liver. UGT1A9 was resequenced up to –5.5 kilobase (kb) of the ATG and included intronic sequences. UGT1A1 and UGT1A9 protein content was measured to identify polymorphisms and haplotypes/diplotypes that might influence gene expression and SN-38G formation in liver microsomes.
Materials and Methods
Reagents. SN-38 was prepared by hydrolysis of irinotecan-HCL (McKesson, Mississauga, Canada). Identity of the SN-38 was assessed by mass spectroscopy, and purity was assessed by UV absorbance spectra. Bilirubin, estradiol, and estradiol-3-glucuronide were purchased from Sigma-Aldrich (St. Louis, MO).
Genomic DNA and Liver Samples. Human genomic DNA and liver samples were obtained from different sources as described previously (Court et al., 2002) as approved by the respective human research institutional review boards (CHUQ Research Center, Laval University and Tufts University). Available subject information included gender, age, race, ethnicity, and histories of smoking and alcohol use, as described in Hesse et al. (2004). The quality of the liver samples was ascertained by reference to at least 10 other glucuronidation activities measured using the same set of livers. Livers that consistently showed low activity values (>2-fold lower for all the measured activities) relative to the median activity value for the entire liver set had been excluded from study.
Genotyping ofUGT1A1andUGT1A9Polymorphisms.UGT1A1 –53TA6>7, –3156G>A, and –3279T>G genotypes for all 48 subjects were determined in a previous study by Girard et al. (2005). Genotyping of UGT1A9 exon 1 polymorphisms at codon 3 and 33 and 11 promoter polymorphisms from –87 to –2152 were performed previously (Girard et al., 2004). For the discovery of new variants, primers R503, F506, R507, F838, R839, F840, R841, and F842 (Table 1) were used to amplify the UGT1A9 promoter region located between nucleotides –1654 and –5525 (relative to the ATG). Genotyping of the two new polymorphisms located at positions –4549 and –5366 was performed by direct sequencing. Primers F1159/R1164 and F1154/R1165 were used to amplify regions with polymorphisms –4549 and –5366, respectively. The polymerase chain reaction (PCR) products were submitted to sequencing using primer R1164 and F1154. The UGT1A9 intron polymorphisms at positions 143C>T, 152G>A, 201A>C, 219T>A, 313A>C, and the novel 399C>T (relative to the end of UGT1A9 exon 1) were genotyped by direct sequencing, with primer F1293, of a PCR product generated with primers F1293 and R1298.
Primers used for amplification, sequencing, and directed mutagenesis
Construction of Plasmid and Transfections. Primers F1121 and R670 were used to amplify the proximal promoter region of UGT1A9 between –152 and +2 (relative to the ATG). The PCR product was digested with HindIII and XhoI and inserted in the pGL3 vector. Primers F1473 and R1474 were also used to amplify the proximal promoter region of UGT1A9 between –171 and –1 (relative to the ATG). The PCR product was digested with SmaI and inserted in the pGL3 vector. A variant sequence was generated by site-directed mutagenesis for both strategies (QuikChange Site-Directed Mutagenesis Kit, Stratagene, La Jolla, CA) with primers F1155 and R1166.
HepG2 cells were obtained from American Type Culture Collection (Manassas, VA) and were grown in minimum essential medium. Medium was supplemented with 20% fetal bovine serum (Wisent, St-Bruno, Canada), as well as a 0.1 mM mixture of nonessential amino acids, 100 IU/ml penicillin, and 50 μg/ml streptomycin (all from Invitrogen, Burlington, Canada). Sodium pyruvate at a final concentration of 1 mM was added. Transfections were performed as described previously (Duguay et al., 2004a). Cells were harvested 24 h post-transfection and assayed for promoter activity using the dual luciferase reporter assay system according to the manufacturer's recommendations, including as an internal control, the pRLnull vector (encoding Renilla luciferase) cotransfected in each well (Promega, Madison, WI). Luciferase activity was measured using 40 μl of cell lysates in a 96-well plate on a microplate luminometer LB96V (EG&G Berthold, Bad Wildbad, Germany) and normalized to the Renilla luciferase values. Means of duplicate luciferase activity of four independent experiments were compared between the different constructs with a two-tailed unpaired Student's t test.
UGT1A1 and UGT1A9 Protein Quantification and Enzymatic Assays. UGT1A1 and UGT1A9 protein quantification in all 48 liver microsomes was performed by semiquantitative Western blot analysis using specific antibodies for UGT1A1 and UGT1A9 as described previously (Duguay et al., 2004b; Girard et al., 2004). The relative levels of UGT protein content were compared with the sample with the lowest expression. SN-38 enzymatic assays were performed in a final volume of 100 μl containing 50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 2 mM UDP-glucuronic acid, 5 μg/ml pepstatin, 0.5 μg/ml leupeptin, 10 μg/ml phosphatidylcholine, 0.2 mg/ml alamethicin (Sigma-Aldrich Chemical Co., Oakville, Canada), 0.5 mM EDTA, 5 μM SN-38, and 8 μg of liver microsomes from each subject as previously described (Gagne et al., 2002). Reactions were stopped by the addition of 100 μl of methanol 2% HCL 2N. SN-38G detection and quantification were performed by liquid chromatography/tandem mass spectrometry as described previously (Gagne et al., 2002; Villeneuve et al., 2003). Estradiol 3-glucuronidation activities were determined as previously described (Krishnaswamy et al., 2003). For bilirubin glucuronidation activities, 100-μl incubations were prepared containing 50 mM phosphate buffer (pH 7.5), 10 μg of liver microsomal protein, 10 μM bilirubin (with 2% dimethyl sulfoxide as a solubility enhancer), 0.5 μg of alamethicin, and 5 mM UDP-glucuronic acid. Because of light sensitivity, bilirubin stock solutions (0.5 mM in dimethyl sulfoxide) were prepared fresh each day and protected from light, and incubations were performed in 0.5-ml amber tubes under minimal light conditions. Incubations were performed for 10 min in a 37°C water bath, terminated by addition of 50 μl of cold acetonitrile containing 5% glacial acetic acid and 5 nmol of 3′-azidothymidine (as internal standard), and then vortexed. After centrifuging for 10 min at 14,000g, the supernatant was transferred to high-performance liquid chromatography (HPLC) tubes, dried down in a vacuum oven set at 45°C, and reconstituted with 100 μl of 0.1% trifluoroacetic acid in water before HPLC analysis. HPLC (Agilent 1100 system, Agilent Technologies, Palo Alto, CA) was performed using a 4.6 × 250-mm, 10-μm C18 column (Synergi Hydro-RP, Phenomenex, Torrance, CA), and effluent was monitored using a UV absorbance diode array detector set at 450 nm (bilirubin and glucuronide) and 254 nm (internal standard). Mobile phase run at 1 ml/min consisted of 0.1% trifluoroacetic acid with 10% acetonitrile in water initially, with a linear gradient to 100% acetonitrile over 20 min, and held at 100% acetonitrile for a further 10 min before returning to initial conditions over 5 min. Internal standard, bilirubin glucuronide, and bilirubin eluted at 8, 15, and 28 min, respectively. Glucuronide peak identity was confirmed by HPLC-mass spectroscopy (ThermoFinnigan DecaXP-Plus, Thermo Electron, Somerset, NJ) and sensitivity to treatment with β-glucuronidase and alkaline pH adjustment. Because bilirubin glucuronide was not available for use as a standard, quantitation was achieved by use of a standard curve generated using known bilirubin concentrations assuming similar UV absorbance of substrate and glucuronide. Results were expressed as nanomole equivalents of bilirubin glucuronide formed per minute per milligram microsomal protein. Preliminary studies confirmed linear metabolite formation for up to 30 min and 0.35 mg/ml microsomal protein concentration. Both within and between-assay variations were less than 15%.
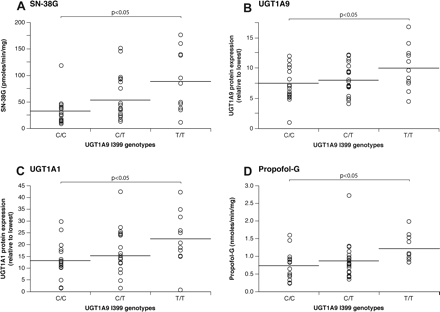
Correlation analyses of UGT1A9 intronic I399 variant genotypes with SN-38 glucuronidation activities (A), UGT1A9 protein expression (B), UGT1A1 protein expression (C), and the UGT1A9-specific substrate propofol (D).
Haplotype and Linkage Analyses. Haplotypes for UGT1A1 and UGT1A9 were determined using Phase v2.1 program (Stephens et al., 2001), and analysis was performed with Caucasian subjects only (n = 42). The linkage between the different polymorphisms was determined with the linkage disequilibrium (LD) plotter tool program found at https://innateimmunity.net/. All the statistical analyses were done using the JMP v4.0.2 program (SAS Institute, Cary, NC) and the SigmaStat program (v3.1; SPSS Inc., Chicago, IL). Correlation analyses between activity values were done using the Spearman's test. R values higher than 0.50 and p values less than 0.05 were considered significant. One-way analysis of variance and a comparison for each pair using Student's t test were used to determine the relationship between genotypes/diplotypes and protein levels or glucuronosyltransferase activities. Only diplotypes with more than three subjects were included in the analysis. The normal distribution of all the expression and activity values was evaluated with the Shapiro-Wilk W test (p > 0.05). Raw data that were not normally distributed were transformed with a logarithm function to achieve a normal distribution that is essential for parametric test. For all the analyses, a p value less than 0.05 was considered significant.
Results
To generate a comprehensive basis for investigating the role of UGT1A9 polymorphisms on SN-38 glucuronidation, we first extended the search for genetic polymorphisms to 5.5 kb of upstream sequence and included intronic sequences. We report two novel promoter single-nucleotide polymorphisms, –5366G>T and –4549T>C (Fig. 1), in strong LD with the previously reported functional variations –2152C>T (LD, r2 = 1.0) and –275T>A (LD, r2 = 0.65). All four polymorphisms are associated with significantly higher hepatic UGT1A9 protein expression in heterozygous subjects (1.6-fold) (Table 2). The previously reported functional UGT1A9 –118T9/10 variant (Yamanaka et al., 2004) was also observed at a frequency of 0.4. Intronic variants recently identified at positions I152A, I219A, and I313C (Carlini et al., 2005) were found in the population tested at an allelic frequency between 0.13 and 0.4, whereas the I143T and I201C were not observed. A novel UGT1A9 intronic variant at position I399C>T was found at a frequency of 0.44.
UGT1A1 and UGT1A9 protein expression and glucuronosyltransferase activities in human livers genotyped for functional polymorphisms
Data are expressed as mean ± S.D.
Correlation analyses were then performed with enzymatic activities, UGT protein content in livers, and UGT1A9 polymorphisms. Compared with individuals with the UGT1A9 I399C/C genotype, subjects with the UGT1A9 I399T/T genotype had a significant increase in SN-38G formation (p < 0.05) (Fig. 2A). In addition, UGT1A1 and UGT1A9 hepatic protein content was elevated in this group of subjects (p < 0.05) (Fig. 2, B and C). This elevation in UGT protein content was reflected at the level of glucuronidation activities. Glucuronidation of UGT1A1 substrates, namely, bilirubin and estradiol-3, as well as UGT1A9 substrates MPA and propofol (Fig. 2D), were significantly increased in subjects with the UGT1A9 I399T allele (p < 0.05) (Table 2).
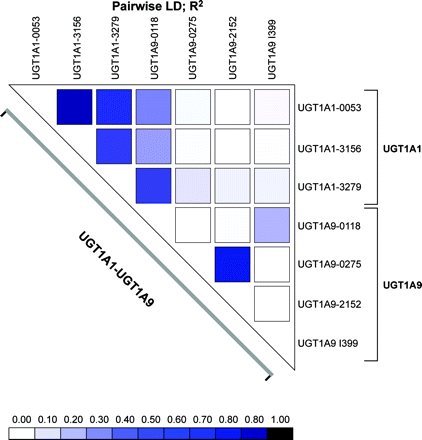
In the population tested, the intronic UGT1A9 I399T variant was not linked to the UGT1A1 –53, –3156, and –3279 variants (r2 < 0.06) (Fig. 3). In addition, a very low degree of LD exists between UGT1A9 I399T and all the other UGT1A9 functional variants (r2 < 0.18) in the subjects tested (Fig. 4). When restricted to subjects with the UGT1A1 –53(TA)6/6 genotype, individuals with the UGT1A9 I399T/T genotype presented 1.7-fold higher UGT1A1 protein content and 2.5-fold higher SN-38 glucuronidation activity compared with UGT1A9 I399C/C carriers (p < 0.05) (Fig. 4). Because of the limited sample size, we could not verify the effect of the UGT1A9 I399 genotype within homozygous subjects for the UGT1A1 –53(TA)7/7 genotype. Similar results were observed for bilirubin and estradiol glucuronide levels (1.8-fold and 2.0-fold; p < 0.05; data not shown). These observations suggest that the UGT1A9 I399T variant may be functional or linked to an unknown functional variant in the UGT1 gene affecting SN-38 glucuronidation levels.
Haplotype structure of UGT1A9 determined with the Phase program in Caucasian subjects only (n = 42). Haplotypes with at least one genotype with less than 90% confidence were excluded. *, represents previously unreported polymorphisms.
The UGT1A9 –118T10 variant was also significantly associated with higher rates of SN-38G formation (p = 0.05) (Table 2 and Fig. 5A). Despite the fact that this polymorphism was not significantly associated with variation of hepatic UGT1A9 protein content (p > 0.05), homozygous –118T10/T10 individuals showed significantly higher levels of UGT1A1 protein compared with individuals –118T9/T9 (p < 0.05) (Fig. 5C). Consistent with this observation, the –118T10 variant was associated with higher glucuronidation of UGT1A1 substrates estradiol and bilirubin (p < 0.05) (Table 2). Conversely, the UGT1A9 –118T10 was not associated with significant alteration of the glucuronidation of UGT1A9-specific substrates (propofol and MPA) (Girard et al., 2004), suggesting an indirect effect of this variation on UGT1A1 expression and UGT1A1-mediated glucuronidation activity. To further evaluate this hypothesis, HepG2 cells derived from human liver cells were transfected with various constructs of the UGT1A9 promoter containing 9 or 10 T repeats at position –118. Results revealed a modest functional impact of 1.4-fold higher expression associated with the –118T10 compared with the –118T9 (p = 0.004) (Fig. 5D). This modest increase may explain the lack of significant alteration of the UGT1A9 protein content and UGT1A9-mediated glucuronosyltransferase activities in liver microsomes.
Pairwise LD r2 for the UGT1A1 and UGT1A9. The LD was calculated with the LD plotter program. The color tone indicates the extent of LD between the two loci.
Correlation analyses of UGT1A9 –118Tn genotypes with SN-38 glucuronidation activities (A), UGT1A9 protein expression (B), and UGT1A1 protein expression (C). The horizontal bars indicate mean values. D, relative luciferase activities for pGL3, UGT1A9 T9, and UGT1A9 T10 constructs in HepG2 cell line are illustrated (*, p = 0.004).
In the population studied and when restricted to Caucasian subjects (n = 42), a low degree of LD was observed between the UGT1A9 I399C>T and the –118T9/10 variant (r2 = 0.19). In contrast, the UGT1A9 –118T10 was found to be linked to the –3279G variant of UGT1A1 (r2 = 0.40) (Fig. 3). This linkage may explain part of the effect of the UGT1A9 –118T10 allele on UGT1A1 protein and activity levels. Of interest, LD was relatively low between UGT1A1 –53 and UGT1A9 –118T (r2 = 0.26).
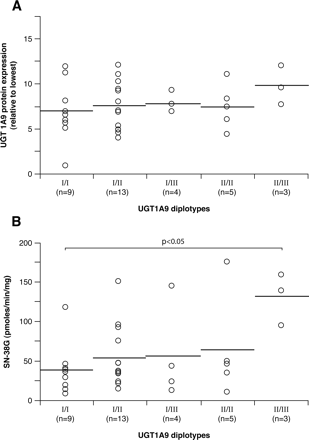
Figure 6 shows the results of UGT1A9 diplotype analyses, restricted to polymorphisms significantly associated with altered protein expression or glucuronosyltransferase activities (–5366, –4549, –2152, –275, –118, and I399). The haplotype UGT1A9_I (GTCT9C) is the most frequent (0.46) and corresponds to the reference UGT1 sequence (AF297093) (Fig. 1). Haplotype UGT1A9_II (GTCT10T), with the variant –118T10 and –I399T, is the second most frequent haplotype (0.33). Haplotype UGT1A9_III (GTCT9T), characterized I399T, is also common in the Caucasian population (0.13). Three other haplotypes (UGT1A9_IV, V, and VI) were found but at relatively low frequencies (0.05, 0.03, and 0.01). There was no significant association between UGT1A9 diplotypes and UGT1A9 hepatic protein content (Fig. 6A). However, compared with subjects with the UGT1A9_I × UGT1A9_I reference diplotype, subjects with the UGT1A9_II × UGT1A9_III diplotype presented higher SN-38 glucuronidation activities (p < 0.05) (Fig. 6B). The impact of the haplotypes characterized by the presence of the –5366T, –4549C, –2152T, and –275A polymorphisms could not be assessed because of their low frequencies.
SN-38 glucuronidation activities in subjects with the UGT1A1 –53(TA)6/6 genotype stratified by UGT1A9 I399C>T genotypes. The horizontal bars indicate mean values.
UGT1A9 protein expression (A) and SN-38 glucuronidation activities (B) associated with UGT1A9 diplotypes, determined by Phase program analysis in Caucasian subjects only. The horizontal bars indicate mean values. N corresponds to the number of subjects in this group. Only diplotypes having n ≥ 3 measurements are reported. Significant results are shown.
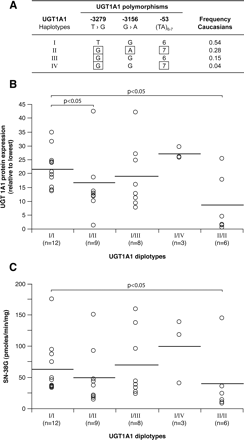
We then evaluated the impact of the UGT1A1 diplotypes (Fig. 7). First, we observed that the UGT1A1 variants –53 (UGT1A1*28, TA7), –3156G>A, and –3279T>G were individually associated with lower UGT1A1 protein content and decreased glucuronidation activities for SN-38 and for UGT1A1 probe substrates bilirubin and estradiol (Table 2). Compared with diplotype UGT1A1_I (TG6) × UGT1A1_I, a significant association of diplotype UGT1A1_II (GA7) × UGT1A1_II with UGT1A1 protein content and SN-38 glucuronidation was shown, consistent with previous reports (p < 0.05) (Fig. 7, B and C) (Innocenti et al., 2004). This relationship was also significant for bilirubin and estradiol (data not shown). A gene-dosage effect could be appreciated for UGT1A1 protein content and SN-38G with an intermediate level of protein or glucuronide for livers with diplotype UGT1A1_I × UGT1A1_II.
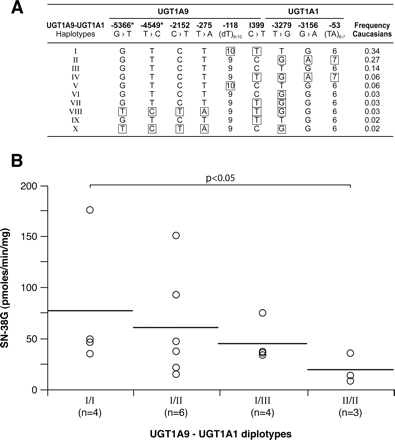
Finally, although our samples size was limited, these results led us to explore the haplotype structure of UGT1A1 and UGT1A9 variants in the Caucasian subjects tested. Ten unambiguous UGT1A1-1A9 haplotypes were inferred. Livers with diplotype UGT1A1-1A9_II (GTCT9CGA7) × UGT1A1-1A9_II had a lower, but not significant, UGT1A1 protein level and significantly lower glucuronidation activities for SN-38 (p < 0.05) (Fig. 8), bilirubin, and estradiol (p < 0.05) (data not shown), with intermediate levels for livers with diplotype UGT1A1-1A9_I × UGT1A1-1A9_II. No significant association of diplotypes with UGT1A9 protein content was detected.
Discussion
Findings show that the UGT1A9 –I399 and –118 polymorphisms have an influence on the hepatic glucuronidation of SN-38 in vitro and that their effects are likely indirect or through their linkage to unknown functional UGT1 variant(s).
The presence of a previously unreported variation in the intron 1 of UGT1A9 (I399C>T) is highly predictive of SN-38 glucuronidation rates by human liver microsomes. In fact, this variant was associated with the most significant and most dramatic increase in SN-38G activity (2.64-fold; p = 0.0007) compared with all the other genotypes, including those of the UGT1A1 promoter. The presence of the UGT1A9 I399T allele is also highly predictive of UGT1A1 protein content in the liver, rates of glucuronidation of specific probe substrates of the UGT1A1 protein, in addition to UGT1A9 protein expression and UGT1A9-mediated activities. Furthermore, that would indicate that this variation might be an independent predictor of SN-38 glucuronidation because within subjects with UGT1A1 –53(TA)6/6 genotype, those with the UGT1A9 –I399T/T presented 2.5-fold higher SN-38 glucuronidation activity (p = 0.016) compared with those with the I399C/C. However, because of the limited liver samples in our bank, we were unable to assess this relationship in subjects homozygous UGT1A1 –53(TA)7/7. Besides, these phenotypic effects do not appear to be explained by linkage to other functional variants because LD was very low between the UGT1A9 I399 and the UGT1A1 –53(TA)n, –3156, and –3279 promoter variants (r2 < 0.06) and with other UGT1A9 variants. The direct functional effects of the intronic variant could not be assessed in this study, but findings suggest that it may be functionally important or link to a functional variant(s) likely unknown because of the lack of LD (r2 < 0.19) with known common functional UGT1A1 and UGT1A9 polymorphisms.
Haplotype structure for UGT1A1 (A) determined with the Phase program in Caucasian subjects only (n = 42). Haplotypes with at least one genotype with less than 90% confidence were excluded. UGT1A1 protein expression (B) and SN-38 glucuronidation activities (C) associated with UGT1A1 diplotypes. The horizontal bars indicate mean values. N corresponds to the number of subjects in this group. Only diplotypes having n ≥ 3 measurements are reported. Significant results are shown.
Two novel UGT1A9 polymorphisms in the 5′ regulatory region identified at positions –5366 and –4549 were found and are tightly linked to those previously reported at positions –2152 and –275. In previous in vitro experiments, –2152T and –275A variants were shown to correlate with higher UGT1A9 hepatic protein content and increased in vitro glucuronidation activities for specific UGT1A9 substrates (Girard et al., 2004). In addition, a recent study in renal transplant recipients established the in vivo functional significance of these variants by measuring a reduced MPA exposure in patients carrying these polymorphisms (Kuypers et al., 2005). Despite in vitro and in vivo evidence of their functionality, these genetic variations of the UGT1A9 gene appear to have no significant effect on the hepatic glucuronidation of SN-38. Overall, findings suggest that the modulation of the UGT1A9 protein in the liver would have a limited impact on the glucuronidation of SN-38.
A recent study indicated that the UGT1A9 –118T9/9 genotype is associated with a lower incidence of toxicity and a better response in colorectal cancer patients treated with an irinotecan-based regimen (Carlini et al., 2005). In our study, the UGT1A9 –118T10 allele was associated with elevated SN-38G formation in livers, higher UGT1A1 protein content, and elevated bilirubin and estradiol glucuronidation. However, it still remains unclear whether this polymorphism is functional. First, it was found that the UGT1A9 –118T10 allele is tightly linked to the high activity UGT1A7*1 and UGT1A1*1 alleles (Carlini et al., 2005). In the liver samples from Caucasians, we confirm that the UGT1A9 –118T10 is in strong LD with the UGT1A1*1 allele and associated with the high activity haplotype I of UGT1A1 (haplotype I TG6). This linkage might explain the relationship between the UGT1A9 –118 variant and irinotecan response rather than reflecting a direct effect of the –118T polymorphism on UGT1A9-mediated glucuronidation of SN-38. Consistent with this hypothesis, livers homozygous for the –118T10 allele showed no significant changes in UGT1A9 protein content and UGT1A9-mediated glucuronidation activities compared with those homozygous for the –118T9 allele. In contrast, subjects homozygous for the –118T10 allele presented significantly higher levels of UGT1A1 protein and glucuronidation activities of UGT1A1-specific substrates, sustaining the hypothesis of an indirect effect.
In addition, results of in vitro gene reporter assays in liver cells support a limited impact of the UGT1A9 –118T10 variation on transcriptional gene activity (1.4-fold increase; p = 0.004) that may not be sufficient to influence levels of UGT1A9 protein in the liver. This is in contrast with a previous study reporting a 2.6-fold greater transcriptional activity associated with a UGT1A9 –118T10 promoter construct compared with –118T9 in HepG2 liver cells. These inconsistencies between studies are somehow difficult to explain because the transfected cell line and the UGT1A9 promoter constructs were identical in both studies. Therefore, with the aim to gain further information from this in vitro approach, we subcloned a slightly different promoter region including the UGT1A9 –118 variation and obtained identical results with a 1.4-fold elevation of the reporter gene expression associated with the –118T10 allele. These in vitro observations are in agreement with our previous observation of the lack of association of the presence of the UGT1A9 –118T10 allele with UGT1A9 protein content in liver microsomes (Girard et al., 2004).
When evaluating combined UGT1A1/1A9 haplotypes, we observed a trend toward lower formation of SN-38G in livers with the diplotype I/III (9/10 at –118) compared with those with I/I (10/10 at –118), whereas Innocenti et al. (2005) observed higher, but not significant, SN-38G formation in livers with the UGT1A1/1A9 haplotype (diplotype I/III) compared with those with diplotype I/I. In both studies, the sample size was rather limited to draw definitive conclusions. Nevertheless, overall in vitro data would point to a limited involvement of this variant in the control of UGT1A9 gene expression and raise the possibility of an effect on SN-38 glucuronidation through linkage with UGT1A1 variants or to unknown polymorphism(s). However, this deserves to be verified because the experimental data do not currently support this hypothesis.
Haplotype structure of UGT1A9-UGT1A1 (A) determined with the Phase program in Caucasian subjects only (n = 42). Haplotypes with at least one genotype with less than 90% confidence were excluded. *, represents previously unreported polymorphisms. SN-38 glucuronidation activities (B) associated with UGT1A9-UGT1A1 diplotypes. The horizontal bars indicate mean values. N corresponds to the number of subjects in this group. Only diplotypes having n ≥ 3 measurements are reported. Significant results are shown.
A lack of LD between UGT1A1 –53(TA)n and UGT1A9 –118Tn (r2 = 0.26) and with the UGT1A9 I399 (r2 = 0.05) was observed in Caucasians. This is consistent with results of a recent study by Innocenti et al. (2005). In this study, the LD between the UGT1A1 –3279 and UGT1A9 –118Tn was higher (r2 = 0.39). When considering additional variants such as the UGT1A9 –331/–440, a strong LD (r2 = 0.79) is observed between UGT1A9 and UGT1A1 [–53(TA)n]. This indicates that linkage among the common UGT1 variants extends from UGT1A1 through to at least UGT1A9, corresponding to a genetic distance of about 88 kb. A significant correlation between expression of both proteins in the liver was observed (Rs = 0.48; p = 0.0006). We also observed that among the common UGT1A1/1A9 haplotypes, polymorphisms associated with lower levels of UGT1A1 and UGT1A9 proteins appear on the same haplotype, indicating that subjects with low levels of UGT1A9 are also susceptible to lower expression of UGT1A1.
Clearly, our genotyping results in limited human livers are exploratory in nature; therefore, our results should be interpreted with caution while awaiting replication and further investigation through clinical study. Nevertheless, this study indicates that the novel intronic UGT1A9 I399 and the –118T promoter variation may represent additional candidates in combination with UGT1A1 promoter haplotypes to predict SN-38 glucuronidation profile in vivo.
Acknowledgments
H.G. is a recipient of a studentship award from the Canadian Institutes of Health Research (CIHR) and Canada's Research-Based Pharmaceutical Companies/CIHR. C.G. is holder of the Canada Research Chair in Pharmacogenomics.
Footnotes
-
This work was supported by the Canadian Institutes of Health Research (CIHR; MOP-42392) and Canada Research Chair Program (C.G.), and by Grants GM-61834, GM-74369, DA-05258, MH-58435, DA-13209, DK-58496, DA-13834, AG-17880, and RR-00054 from the National Institutes of Health (M.H.C, D.J.G., L.L.v.M.).
-
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
-
doi:10.1124/dmd.106.009787.
-
ABBREVIATIONS: SN-38, 7-ethyl-10-hydroxycamptothecin; UGT, UDP-glucuronosyltransferase; SN-38G, SN-38-glucuronide; PCR, polymerase chain reaction; HPLC, high-performance liquid chromatography; LD, linkage disequilibrium; MPA, mycophenolic acid.
- Received February 18, 2006.
- Accepted March 29, 2006.
- The American Society for Pharmacology and Experimental Therapeutics