Abstract
UDP-glucuronosyltransferases (UGTs) catalyze glucuronidation of a variety of xenobiotics and endobiotics. UGTs are divided into two families, UGT1 and UGT2. The purpose of this study was to estimate the absolute expression levels of each UGT isoform in human liver and to evaluate the interindividual variability. Real-time reverse transcriptase-polymerase chain reaction analysis was performed to determine the copy numbers of nine functional UGT1A isoforms and seven UGT2B isoforms. We noticed that not only primers but also templates as a standard for quantification should prudently be selected. Once we established appropriate conditions, the mRNA levels of each UGT isoform in 25 individual human livers were determined. UGT1A1 (0.9–138.5), UGT1A3 (0.1–66.6), UGT1A4 (0.1–143.3), UGT1A6 (1.0–70.4), UGT1A9 (0.3–132.4), UGT2B4 (0.3–615.0), UGT2B7 (0.2–97.4), UGT2B10 (0.7–253.2), UGT2B15 (0.3–107.8), and UGT2B17 (0.5–157.1) were substantially expressed (×104 copy/μg RNA) with large interindividual variability. Abundant isoforms were UGT2B4 and UGT2B10, followed by UGT1A1, UGT2B15, and UGT1A6. The sum of the UGT2B mRNA levels was higher than that of UGT1A mRNA levels. It is interesting to note that the mRNA levels normalized with glyceraldehyde-3-phosphate dehydrogenase mRNA for almost UGT isoforms that are substantially expressed in liver showed significant correlations to each other. Western blot analysis was performed using antibodies specific for UGT1A1, UGT1A4, UGT1A6, or UGT2B7. Correlation between the protein and mRNA levels was observed in only UGT1A1 (r = 0.488; p < 0.01). In conclusion, this study comprehensively determined the absolute values of mRNA expression of each UGT isoform in human livers and found considerable interindividual variability.
UDP-glucuronosyltransferase (UGT) enzymes catalyze glucuronidation of a variety of xenobiotics and endogenous compounds (Tukey and Strassburg, 2000). In humans, UGTs are classified into UGT1 and UGT2 families, and the latter is divided into UGT2A and UGT2B subfamilies based on evolutionary divergence and homology (Mackenzie et al., 2005). The human UGT1A gene complex is located on chromosome 2q37 and consisted of multiple unique first exons and common exon 2 to 5 (Gong et al., 2001), encoding nine functional members of the UGT1A subfamily. The UGT2 gene family is located on chromosome 4q13 and includes three members of the UGT2A subfamily and seven functional members of the UGT2B subfamily. Each UGT2 gene comprises six exons that are not shared between the UGT2 family members, with an exception of UGT2A1 and UGT2A2, which are arisen by the differential splicing of a variable first exon to the same set of five downstream exons, similar to the UGT1A enzymes (Mackenzie et al., 2005).
Liver is the major organ for glucuronidation in the body as it is directly exposed to the influx of drugs from the hepatic portal vein during oral absorption. Earlier studies showed that the mRNAs of all the UGT isoforms except for UGT2A1 were expressed in the human liver at any level (Strassburg et al., 1997a; Tukey and Strassburg, 2000, 2001; Fisher et al., 2001; Aueviriyavit et al., 2007; Court et al., 2008; Nakamura et al., 2008b). In most studies, the mRNA expressions were qualitatively evaluated using reverse transcriptase-polymerase chain reaction (RT-PCR). Recently, the UGT expression levels have been quantitatively determined using real-time RT-PCR, but the data have been evaluated as relative values (Aueviriyavit et al., 2007; Court et al., 2008). Therefore, we could not directly compare the expression levels of different UGT isoforms. If we could know the absolute expression levels of each UGT isoform, the information might be useful to estimate the contribution of each UGT isoform in certain glucuronidation that is catalyzed by multiple UGT isoforms. Moreover, there are limited data on the interindividual variability in UGT expression levels in human livers. Because interindividual variability of UGT expression in livers plays an important role in drug efficacy, toxicity, and susceptibility to environmental chemicals (Wells et al., 2004), the analysis of interindividual variability of each UGT expression is important. The aims of this study were to determine the absolute values of copy numbers of UGT isoforms in human liver and to evaluate their interindividual variability. Although it is possible to quantify UGT protein levels using selective antibodies, immunodetection of UGT proteins is plagued by uncertainty regarding antibody specificity because UGT families possess a high degree of protein sequence homology. Therefore, their use is restricted to the few UGT enzymes for which selective antibodies exist. Another aim of this study was to investigate the relationship between mRNA levels and protein levels for certain UGT isoforms.
Materials and Methods
Materials. RNAiso, random hexamer, and SYBR Premix Ex Taq were from Takara Bio (Shiga, Japan). ROX was purchased from Stratagene (La Jolla, CA). ReverTra Ace (Moloney murine leukemia virus reverse transcriptase RNaseH–) was obtained from Toyobo (Tokyo, Japan). Primers were commercially synthesized at Hokkaido System Sciences (Sapporo, Japan). Rabbit anti-human UGT1A1 polyclonal antibody and anti-human UGT2B7 polyclonal antibody were purchased from BD Gentest (Woburn, MA). Rabbit anti-human UGT1A4 and UGT1A6 peptide polyclonal antibodies were prepared previously (Ikushiro et al., 2006). All the other reagents were of the highest grade commercially available.
Human Livers. Human liver samples from 16 donors (10 white, 4 Hispanic, 1 black, and 1 Asian) were obtained from Human and Animal Bridging Research Organization (Chiba, Japan), and those from nine Japanese donors were obtained from autopsy materials that were discarded after pathological investigation (Supplemental Table 1). The use of the human livers was approved by the Ethics Committees of Kanazawa University (Kanazawa, Japan) and Iwate Medical University (Morioka, Japan).
Total RNA and Reverse Transcription. Total RNA was extracted from 25 individual human liver samples using RNAiso. The integrity of the RNA was assessed by estimating the ratio of 28S and 18S rRNA bands on ethidium bromide–stained 1% agarose gel. The cDNA was synthesized using ReverTra Ace according to the manufacturer's protocols.
Isolation and Subcloning of Human UGT cDNAs. Human UGT1A3, UGT1A5, UGT1A7, UGT1A8, UGT1A10, UGT2B4, UGT2B7, UGT2B10, UGT2B11, UGT2B15, UGT2B17, and UGT2B28 cDNAs were prepared by RT-PCR using total RNA from appropriate human tissues or various cell lines (Nakamura et al., 2008b). The used primers are shown in Table 1. After an initial denaturation at 94°C for 5 min, amplification was performed by denaturation at 94°C for 25 s, annealing at an appropriate temperature for 25 s, and extension at 72°C for 2 min for 40 cycles. The final extension step was performed at 72°C for 5 min. These PCR products were subcloned into pTARGET Mammalian Expression Vector. The plasmid DNA was purified by a QIAGEN Plasmid Midi kit (QIAGEN, Valencia, CA) and submitted to DNA sequences using a Thermo Sequenase Cy5.5 Dye Terminator Cycle Sequencing kit (GE Healthcare, Little Chalfont, Buckinghamshire, UK) with a Long-Read Tower DNA sequencer (GE Healthcare). The pTARGET plasmids containing human UGT1A1, UGT1A4, UGT1A6, and UGT1A9 were previously constructed (Fujiwara et al., 2007). These plasmids were digested with appropriate restriction enzymes to prepare the standards for real-time RT-PCR analysis. As a standard for real-time RT-PCR analysis, amplification efficiencies with the intact plasmid, linearized plasmid, full-length cDNA, and the purified product of the real-time RT-PCR per se were compared.
Sequence of primers used for amplification of UGT cDNAs
Preliminary PCR Analyses to Validate the Amplification with Different Primer Pairs. A1-μl portion of the reverse-transcribed mixture was added to PCR mixtures (25 μl) consisting of 1× PCR buffer (67 mM Tris-HCl buffer, pH 8.8, 16.6 mM (NH4)2SO4, 0.45% Triton X-100, 0.02% gelatin), 1.5 mM MgCl2, 0.4 μM primers, 250 μM dNTPs, and 1 U of Taq DNA polymerase (Greiner Japan, Tokyo, Japan). The primer pairs are 1A4 ex1-S and 1A4 ex1-AS, int-AS, or 1A ex2-AS; 1A5 ex1-S and 1A5 ex1-AS, int-AS, or 1A ex2-AS; and int-S and 1A ex2-AS. The sequences of the primers are shown in Table 2. PCR reaction was performed with Takara PCR Thermal Cycler Dice TP600 (Takara Bio). After an initial denaturation at 94°C for 5 min, amplification was performed by denaturation at 94°C for 25 s, annealing at 58°C for 25 s, and extension at 72°C for 40 s for 25 cycles. The final extension step was performed at 72°C for 5 min. The PCR products (8 μl) were analyzed by electrophoresis with 2% agarose gel and visualized by ethidium bromide staining.
Sequence of primers used for RT-PCR analyses
Real-Time RT-PCR Analysis. The UGT mRNA levels in human livers were quantified by real-time RT-PCR using the Mx3000P (Stratagene). PCR mixture contained a 1-μl portion of the reverse-transcribed mixture, SYBR Premix Ex Taq solution, and 0.4 μM primers. The sequences and position of the primers, as well as annealing temperatures, are shown in Table 3. After an initial denaturation at 95°C for 30 s, amplification was performed by denaturation at 94°C for 4 s, annealing at an appropriate temperature for 7 s, and extension at 72°C for 20 s for 45 cycles. Amplified products were monitored directly by measuring the increase of the dye intensity of the SYBR Green (Takara Bio) that binds to double-strand DNA amplified by PCR. Copy number in the samples was defined based on a standard curve using full-length UGT cDNA. The specificity of all the primer pairs was confirmed by digestion of the PCR products with appropriate restriction enzymes and sequence analysis. Negative control samples were processed in the same manner, except that the template was omitted. A calibration curve was constructed by plotting the PCR threshold cycle (Ct) number at which the fluorescent signal generated during the replication process passes above a threshold value against known amounts of cDNA. UGT mRNA expression levels were normalized with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA level.
Sequence of primers used for real-time RT-PCR analyses
Western Blot Analysis. Microsomes from 25 human livers were prepared according to a method described previously (Tabata et al., 2004). The human liver microsomes (10 μg) were separated on 10% SDS-polyacrylamide gel and transferred electrophoretically to either polyvinylidene difluoride or nitrocellulose membrane. Polyvinylidene difluoride membrane Immobilon-P (Millipore Corporation, Billerica, MA) was probed with anti-human UGT1A1 antibody or anti-human UGT2B7 antibody. In the data sheet provided by the manufacturer, it is described that the UGT1A1 antibody did not cross-react with UGT1A4, UGT1A6, UGT1A9, UGT1A10, and UGT2B15 and that the UGT2B7 antibody did not cross-react with UGT1A1, UGT1A4, UGT1A6, UGT1A9, UGT1A10, and UGT2B15. We confirmed that UGT2B7 antibody did not cross-react with UGT2B4, UGT2B15, and UGT2B17 using the recombinant enzymes expressed in baculovirus-infected insect cells (Supersomes) (data not shown). Nitrocellulose membrane (Whatman Biometra GmbH, Niedersachsen, Germany) was probed with anti-human UGT1A4 antibody or antihuman UGT1A6 antibody. These antibodies have shown no cross-reactivity with the other UGT1A isoforms (Ikushiro et al., 2006). Biotinylated anti-rabbit IgG and a Vectastain ABC kit (Vector Laboratories, Burlingame, CA) were used for diaminobenzidine staining. The expression level of UGT protein was defined based on a standard curve using recombinant human UGTs expressed in baculovirus-infected insect cells (BD Gentest). The quantitative analysis was performed using a GT-9800F scanner (Seiko Epson, Suwa, Japan) and ImageQuant TL Image Analysis software (GE Healthcare).
Preliminary RT-PCR analyses to determine the specificity targeting UGT1A4 or UGT1A5 with different primer sets. RT-PCR was performed using the sense and antisense primers that are in the exon 1 of UGT1A4 or UGT1A5 (A). RT-PCR was performed using the sense primer in exon 1 and antisense primer in intron 1 of UGT1A4 or UGT1A5 (B). RT-PCR was performed using the sense primer in intron 1 of UGT1A4 or UGT1A5 and antisense primer in exon 2 (C). RT-PCR was performed using the sense primer in exon 1 of UGT1A4 or UGT1A5 and antisense primer in exon 2 (D). The numbers under each photograph represent the sample numbers. Schematic representation of UGT1A gene, pre-mRNA, mRNA, and excised intron (E). Boxes represent exons, and lines represent introns. The location and direction of primers were shown as arrows. PCR was performed with various primer sets.
Statistical Analysis. Data are expressed as the mean ± S.D. Correlation analysis was performed by Spearman rank method. A value of p < 0.05 was considered statistically significant.
Results
Primer Design to Quantify the UGT mRNA. For the specific amplification of each human UGT1A mRNA, many investigators used sense and antisense primers that are within exon 1 (Congiu et al., 2002; Finel et al., 2005; Kaivosaari et al., 2007; Itäaho et al., 2009; Ohno and Nakajin, 2009) because the exon 2 to 5 is common to all the UGT1A isoforms. To avoid overestimation by contamination of genomic DNA, the sense and antisense primers are preferable to be located in different exons. In this study, we compared the amplification with different primer sets for UGT1As, targeting UGT1A4 and UGT1A5, which are highly and marginally expressed in liver, respectively. When the primer pairs were located in exon 1 of UGT1A4 (UGT1A4 ex1-S and UGT1A4 ex1-AS) or in exon 1 of UGT1A5 (UGT1A5 ex1-S and UGT1A5 ex1-AS), PCR amplicon was detected with the reverse-transcribed mixture but not with the mixture excluding reverse transcriptase (Fig. 1A), suggesting that the contamination of genomic DNA was negligible. Nevertheless, when the sense and antisense primers were located in exon 1 and intron 1, respectively, for UGT1A4 (UGT1A4 ex1-S and int-AS) and UGT1A5 (UGT1A5 ex1-S and int-AS), obvious amplicon was detected (Fig. 1B), indicating the presence of pre-mRNA or excised intron in a lariat shape (Fig. 1E). Because the primer set of int-S and UGT1A ex2-AS also show an obvious band (Fig. 1C), the presence of pre-mRNA was supported. This would not be specific for the UGT1A family because the primer sets located in exon 1 and intron 1 for UGT2B7, UGT2B10, and UGT2B28 showed obvious amplification with the reverse-transcribed mixture (data not shown). Collectively, if the primers were set in the same exon, the amplification of not only genomic DNA but also pre-mRNA would occur. The sense and antisense primers should properly be in different exons (Fig. 1D).
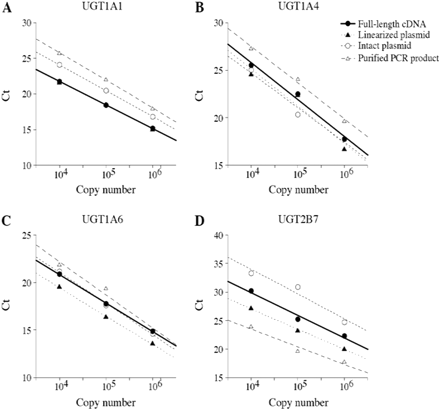
Standard Curves Using Different Length of cDNA Fragments as a Template. Plasmids containing full-length cDNA are frequently used as a template to calculate the copy numbers in RNA samples in real-time RT-PCR analysis. We examined the extent of amplification with the intact plasmid, the linearized plasmid, and the purified PCR product by the real-time RT-PCR compared with full-length cDNA. Figure 2 shows Ct number with 104, 105, or 106 copies of these templates. In the case of UGT1A1, the standard curve with the linearized plasmid was overlaid with that of full-length cDNA. In contrast, Ct values with the intact plasmid or PCR product were greater than those with full-length cDNA (Fig. 2A). In the case of UGT1A4, the standard curves with the intact plasmid and linearized plasmid were shifted to lower Ct values, but that with the PCR product was shifted to higher Ct values, comparing with that of full-length cDNA (Fig. 2B). In the case of UGT1A6, the standard curve with the intact plasmid was overlaid with that of full-length cDNA (Fig. 2C). In contrast, the Ct values with the linearized plasmid were lower than those with full-length cDNA. The Ct values with the PCR product were higher than those with full-length cDNA. In the case of UGT2B7, four kinds of standard curves were not overlaid at all (Fig. 2D). Taken together, these results suggest that the absolute values of copy number would be misjudged depending on the kinds of template. For the subsequent study, the full-length cDNAs were used as a template for the quantification because it is exactly the reverse-transcribed product of intact mRNA. We confirmed that freezing and thawing of the full-length cDNAs did not affect the quantification (data not shown).
The standard curves for UGTs by real-time RT-PCR. The intact or linearized plasmid containing cDNA, full-length cDNA, and purified PCR product for UGT1A1 (A), UGT1A4 (B), UGT1A6 (C), and UGT2B7 (D) was used as the template. The standard curves were generated by plotting the Ct of the crossing points versus the copy number of template.
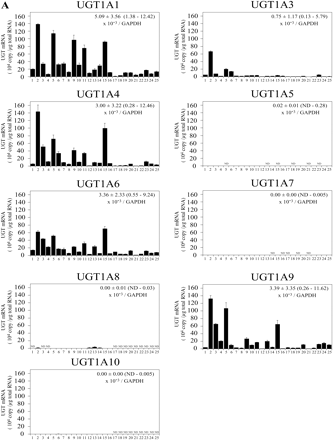
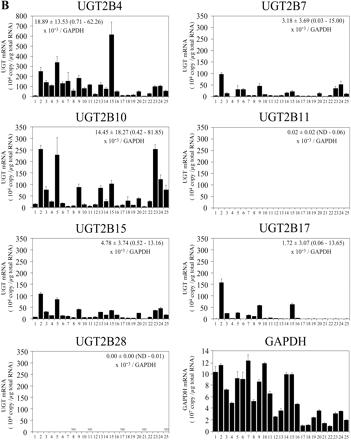
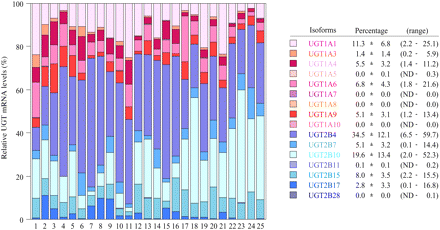
Expression Level of UGT mRNA in Human Livers. The expression levels of UGT1A and UGT2B mRNA in 25 human livers were determined by real-time RT-PCR (Fig. 3). UGT1A1 mRNA levels were 0.9 to 138.5 × 104 copy/μg. UGT1A3 mRNA levels were 0.1 to 66.6 × 104 copy/μg. UGT1A4 mRNA levels were 0.1 to 143.3 × 104 copy/μg. UGT1A6 mRNA levels were 1.0 to 70.4 × 104 copy/μg. UGT1A9 mRNA levels were 0.3 to 132.4 × 104 copy/μg. UGT1A5, UGT1A7, UGT1A8, and UGT1A10 mRNAs were marginally expressed. UGT2B4 mRNA levels were 0.3 to 615.0 × 104 copy/μg. UGT2B7 mRNA levels were 0.2 to 97.4 × 104 copy/μg. UGT2B10 mRNA levels were 0.7 to 253.2 × 104 copy/μg. UGT2B15 mRNA levels were 0.3 to 107.8 × 104 copy/μg. UGT2B17 mRNA levels were 0.5 to 157.1 × 104 copy/μg. UGT2B11 and UGT2B28 mRNAs were marginally expressed. After the normalization with GAPDH mRNA levels (0.9 to 12.3 × 107 copy/μg), the interindividual variation was estimated as follows: UGT1A1, 9-fold; UGT1A3, 37-fold; UGT1A4, 28-fold; UGT1A6, 22-fold; UGT1A9, 45-fold; UGT2B4, 72-fold; UGT2B7, 506-fold; UGT2B10, 223-fold; UGT2B15, 29-fold; and UGT2B17, 223-fold. Thus, the UGT2B subfamily showed relatively larger interindividual variability than the UGT1A subfamily. Figure 4 shows the percentage of each UGT mRNA level in sum of all the UGT levels. Abundant isoforms were UGT2B4 (34.5 ± 12.1%, 6.5–59.7% of total UGT) and UGT2B10 (19.6 ± 13.4%, 2.0–52.3%), followed by UGT1A1 (11.3 ± 6.8%, 2.2–25.1%), UGT2B15 (8.0 ± 3.5%, 2.2–15.5%), and UGT1A6 (6.8 ± 4.3%, 1.8–21.6%). The sum of the UGT2B mRNA levels (70.1 ± 11.4%, 42.7–90.0%) was higher than that of UGT1A mRNA levels (29.9 ± 11.4%, 10.0–57.3%).
Expression Level of UGT Protein in Human Livers and Relationship with the mRNA Levels. The expression levels of UGT1A1, UGT1A4, UGT1A6, and UGT2B7 protein in 25 human liver microsomes were determined by Western blot analysis (Fig. 5). Although the specific antibody against UGT1A9 is available (Ikushiro et al., 2006), it showed a large number of nonspecific bands with the human liver microsomes (data not shown). Other antibodies against UGT1A9 obtained from Abnova (Taipei City, Taiwan) cross-reacted with other UGT1As. Therefore, we could not determine the UGT1A9 levels in human liver microsomes. The interindividual variabilities of UGT1A1, UGT1A4, UGT1A6, and UGT2B7 proteins were 6-, 13-, 2-, and 4-fold, respectively. The UGT1A1 protein levels were significantly correlated with the UGT1A1 mRNA levels (r = 0.488; p < 0.01). However, no correlation between the protein and mRNA levels was observed for UGT1A4, UGT1A6, and UGT2B7.
Correlation Analyses of the Expression Levels of UGT mRNA or Protein in Human Livers between Different Isoforms. As for UGT isoforms that are substantially expressed in human livers, correlation analyses of the expression levels of UGT mRNA between isoforms were performed. UGT mRNA levels normalized with GAPDH mRNA in 25 human livers were used for the analysis. We were surprised to find that most isoforms showed significant correlations each other (Table 4). An exception was UGT2B17, which was hardly correlated with other isoforms. In contrast to the results of mRNA levels, at the protein levels, a significant correlation was observed only between UGT1A4 and UGT1A6 (r = 0.441; p < 0.05).
Correlations of the expression levels of UGT mRNA or protein between different isoforms in 25 human livers
Top half shows the correlation between expression levels of UGT mRNAs, and bottom half shows the correlation between expression levels of UGT protein levels. The correlation analyses were performed for UGT isoforms that are substantially expressed in human livers by the Spearman rank method. The Spearman correlation coefficients (r) are shown.
Discussion
In this study, we found that UGT1A1, UGT1A3, UGT1A4, UGT1A6, UGT1A9, UGT2B4, UGT2B7, UGT2B10, UGT2B15, and UGT2B17 were substantially expressed in human liver in agreement with previous studies (Strassburg et al., 1997b, 2000; Tukey and Strassburg, 2001). In addition, UGT1A5, UGT1A7, UGT1A8, UGT1A10, and UGT2B11 were detected in agreement with previous studies (Beaulieu et al., 1998; Zheng et al., 2002; Finel et al., 2005; Nakamura et al., 2008b), although their levels were extremely low. We also examined the expression of UGT2A1, UGT2A2, and UGT2A3 in human livers. Although UGT2A2 and UGT2A3 were detected (data not shown), their Ct values were as high as those for latter isoforms such as UGT1A5. Therefore, we did not determine the copy numbers of UGT2As.
One of the advantages of our study was that the UGT mRNA levels were quantitatively assessed, which allow us to compare directly the expression levels between different isoforms. We found that the abundant isoforms were UGT2B4 and UGT2B10, followed by UGT1A1, UGT2B15, and UGT1A6. In brief, our results were compatible with recent works reporting the quantitative data of UGT mRNA levels in human liver, although they used pooled samples (Kaivosaari et al., 2007; Itäaho et al., 2009; Ohno and Nakajin, 2009). Kaivosaari et al. (2007) and Itäaho et al. (2009) reported using pooled human liver from 47 donors, and the expression levels (copies/109 copies 18S rRNA) are as follows: UGT1A9 (3239 ± 42, mean ± S.E.) > UGT2B10 level (1980, mean) ≈ UGT1A4 (1321, mean) ≈ UGT2B7 (1309, mean) ⋙ UGT1A10 (6 ± 0, mean ± S.E.). Ohno and Nakajin (2009) reported using pooled human liver from three donors, and the abundant isoforms (copies/104 copies GAPDH) are as follows: UGT2B4 (37,900 ± 711, mean ± S.D.) > UGT2B15 (18,500 ± 285) > UGT2B7 (4220 ± 13) > UGT2B10 (3380 ± 93). In dissecting the relative expression levels of UGTs, however, there is discordance between the previous studies and our own. One possibility of the inconsistency might be because of the interindividual variability. Another possibility is the differences in primers and the template used to construct the calibration curve. In the previous studies, sense and antisense primers were both localized in the same exon, and intact plasmids containing cDNA or purified PCR product were used as the template. Earlier studies underappreciated the selection of template, but the present study clearly showed that the different length templates lead to different results. The full-length cDNA would be preferable as the template because it is the reverse-transcribed product of intact mRNA. In the last, we found that the sum of the UGT2B mRNA levels (70.1 ± 11.4%, 42.7–90.0%) was higher than that of UGT1A mRNA levels (29.9 ± 11.4%, 10.0–57.3%).
The expression levels of UGT1A and UGT2B mRNA in 25 individual livers. The expression levels of UGT1A (A) and UGT2B (B) mRNA in human livers were measured by real-time RT-PCR: 1, 2, 6 through 8, 11 through 13, 15, and 16 were from white subjects, 3 through 5 and 10 were from Hispanic subjects, 9 was from a black subject, 14 was from an Asian subject, and 17 through 25 were from Japanese subjects. Each column represents the mean ± S.D. of triplicate data. ND, not detectable.
Another advantage of this study was that the interindividual variability of mRNA levels was assessed for all the UGT isoforms. The interindividual variability of UGT1A mRNA levels was largely consistent with previous studies. Aueviriyavit et al. (2007) have reported that the interindividual variability in 18 human livers were as follows: UGT1A1 (8.6-fold), UGT1A3 (6.5-fold), UGT1A4 (2.5-fold), UGT1A6 (4.9-fold), and UGT1A9 (5.1-fold) mRNA levels. Krishnaswamy et al. (2005) reported that the interindividual variability of UGT1A6 mRNA was 7-fold in 50 human livers. Our study showed that the UGT2B isoforms had a great interindividual variability. Of particular interest is the fact that mRNA levels for most UGT isoforms showed significant correlations each other. Supporting our study, Ramírez et al. (2008) reported that the UGT1A1 mRNA levels were significantly correlated with the UGT1A9 (r2 = 0.49; p < 0.0001; n = 44) and UGT2B7 (r2 = 0.39; p < 0.0001; n = 54) mRNA levels, and that the UGT1A9 mRNA levels were significantly correlated with the UGT2B7 mRNA levels (r2 = 0.54; p < 0.0001; n = 44). They concluded that the results would be because of the common regulation by hepatic nuclear factor (HNF) 1α as these UGT mRNA levels were significantly correlated with the HNF1α mRNA levels. In addition, Aueviriyavit et al. (2007) also reported that UGT1A6 and UGT1A9 mRNA levels were significantly correlated with the HNF1α and HNF4α mRNA levels. In fact, there is literature showing that HNF1α and HNF4α can bind and activate their promoters (Bernard et al., 1999; Ishii et al., 2000; Gregory et al., 2004; Barbier et al., 2005; Gardner-Stephen and Mackenzie, 2007). Thus, these liver-enriched transcriptional factors would be the major determinant of the expression of hepatic UGTs. Besides, hormones, bile acids, drugs, and other xenobiotics may modulate the UGT expression through interaction with receptors such as pregnane X receptor, constitutive androstane receptor, peroxisome proliferator-activated receptor α, farnesoid X receptor, aryl hydrocarbon receptor, and Nrf2 (Zhou et al., 2005; Nakamura et al., 2008a). These factors may also contribute the interindividual variability.
The percentage of mRNA levels of each UGT isoform in sum of all the UGT levels. The mean values of triplicate determinations in Fig. 3 were adopted. The mRNA levels of each UGT isoform normalized with GAPDH mRNA levels were used for this analysis.
Western blot analyses of UGT1A1, UGT1A4, UGT1A6, and UGT2B7 and the relationship between the UGT protein and mRNA levels. Western blot analyses were performed for 25 human liver microsomes using specific antibodies against human UGT1A1, UGT1A4, UGT1A6, and UGT2B7, and typical results are shown for 12 microsomes (A). M, marker. The correlation analyses were performed between the protein and mRNA levels for UGT1A1 (B), UGT1A4 (C), UGT1A6 (D), and UGT2B7 (E) by Spearman's rank method.
Genetic polymorphisms might be another factor determining the variability of UGT expression. UGT1A1*28 possessing TA-inserted promoter (TA)7TA and UGT1A1*60 possessing a single nucleotide polymorphism –3263T>G show reduced transcriptional activity (Guillemette et al., 2001; Sugatani et al., 2002). Two single nucleotide polymorphisms in the promoter region of UGT1A4 gene have been reported to be associated with the reduced transcriptional activity (Erichsen et al., 2008). For the UGT2B17 gene, a polymorphism of the entire gene deletion is known (Wilson et al., 2004; Karypidis et al., 2008). Although we did not investigate whether our samples have such variants, it is possible that the variation in UGT mRNA may partly be explained by the genetic polymorphisms.
We performed Western blot analysis to determine the UGT protein levels in human liver microsomes. Our results showed a significant correlation between mRNA and protein levels for UGT1A1 (r = 0.488; p < 0.01) but not for UGT1A4, UGT1A6, and UGT2B7. Krishnaswamy et al. (2005) previously reported that the UGT1A6 mRNA levels in 54 human livers were significantly (r = 0.53; p < 0.001) correlated with UGT1A6 protein levels. The inconsistency may be because of the differences in the sample numbers and experimental conditions, including antibodies and/or donors. Although we did confirm the linearity of assay response through use of a standard curve constructed using different amounts of recombinant UGTs, the semiquantitative immunoblotting may not adequately show the true range of values. This result might illustrate less correlation between mRNA and protein levels. Alternatively, post-transcriptional and/or post-translational regulation might be feasible, but such mechanisms largely remain to be studied for UGTs. Further studies will be required to draw definitive conclusions.
In conclusion, this is the first study to comprehensively assess the expression levels of each UGT isoform in human livers, evaluating their interindividual variability. The findings presented here provide important insight to understand the interindividual variability of glucuronidations.
Acknowledgments
We thank Brent Bell for review of the manuscript.
Footnotes
-
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
-
doi:10.1124/dmd.109.027227.
-
ABBREVIATIONS: UGT, UDP-glucuronosyltransferase; RT-PCR, reverse transcriptase-polymerase chain reaction; Ct, threshold cycle; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HNF, hepatic nuclear factor.
-
↵
 The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material. - Accepted May 7, 2009.
- Received February 18, 2009.
- The American Society for Pharmacology and Experimental Therapeutics