Abstract
Over the past 20 years, quantitative proteomics has contributed a wealth of protein expression data, which are currently used for a variety of systems pharmacology applications, as a complement or a surrogate for activity of the corresponding proteins. A symposium at the 25th North American International Society for the Study of Xenobiotics meeting, in Boston, in September 2023, was held to explore current and emerging applications of quantitative proteomics in translational pharmacology and strategies for improved integration into model-informed drug development based on practical experience of each of the presenters. A summary of the talks and discussions is presented in this perspective alongside future outlook that was outlined for future meetings.
SIGNIFICANCE STATEMENT This perspective explores current and emerging applications of quantitative proteomics in translational pharmacology and precision medicine and outlines the outlook for improved integration into model-informed drug development.
Introduction
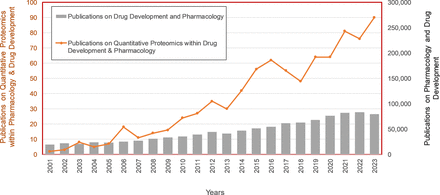
In addition to facilitating drug discovery (Meissner et al., 2022), quantitative proteomics has applications across many translational pharmacology domains, notably in characterizing in vitro systems, assessing interspecies differences, interindividual variability, and developing physiologically based pharmacokinetic (PBPK) models (El-Khateeb et al., 2019; Prasad et al., 2019). The increased application of proteomics in drug development is reflected in Fig. 1, against a generally stable research output in drug development. The use of protein abundance data becomes even more important as the variation of functional activity of proteins associated with a given genotype due to different expression levels are further elucidated, suggesting that pharmacogenomics alone is not a panacea for resolving interindividual variations in drug exposure and response (Polasek, 2024). By analyzing whole proteomes, quantitative proteomics also facilitates discovery and validation of biomarkers for diagnostic and therapeutic applications, enabling precision medicine through patient stratification for tailored treatments (Darwich et al., 2021).
Summary of the number of publications on ‘quantitative proteomics’ in ‘pharmacology’ and ‘drug development’ against a background of publications on ‘pharmacology’ and ‘drug development’ between 2001 and 2023 (database: Web of Science).
The application of quantitative proteomics in drug metabolism and pharmacokinetics (DMPK) research started to rise in 2008 (Kamiie et al., 2008; Li et al., 2009a,b) when the field recognized the utility of the technique for analyzing membrane-anchored and transmembrane proteins that lack antibodies for immunoquantification. A decade of rapid progress in the field with many other groups utilizing the technique for different DMPK applications culminated in a workshop organized in 2018 by the International Society for the Study of Xenobiotics, serving as a platform to deliberate and derive insights into the utilization of quantitative proteomics in DMPK research and precision medicine. The proceedings of this workshop were published in a white paper, summarizing the consensus on methodology and applications of quantitative proteomics in translational pharmacology and precision medicine (Prasad et al., 2019).
Five years after the publication of the white paper (Prasad et al., 2019), a symposium was organized at the 25th North American International Society for the Study of Xenobiotics meeting in Boston in September 2023 to revisit progress made in the field since then. A summary of podium presentations and panel discussions, which revolved around the practical experience of some of the most active groups in the field, is provided below for the benefit of the wider drug development community with interest in this area. Drug Metabolism and Disposition was recognized recently (Wang et al., 2024) as the central journal in a bibliographic analysis of publications informing and applying PBPK. Therefore, the readership of this journal can be considered as the community who might benefit the most from this report.
What is New in Quantitative Proteomics: Techniques and Applications
What is Quantitative Proteomics?
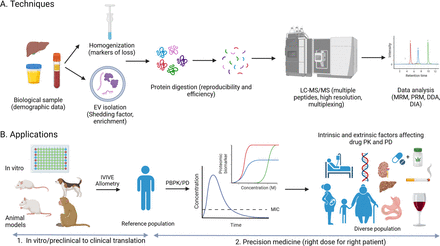
Quantitative proteomics relies on liquid chromatography (LC) and mass spectrometry (MS), enabling multiplexed quantification of proteins that is selective, precise, and in many cases cost-effective. It comprises targeted and untargeted approaches, where targeted proteomics involves the identification of a predetermined set of proteins using selected surrogate peptides (Fig. 2A). Targeted proteomics utilizes (i) triple quadrupole-based tandem mass-spectrometry in multiple or selected reaction monitoring mode or (ii) high-resolution mass spectrometry (HRMS) in parallel reaction monitoring mode, both for precise quantification of specific peptides. For absolute peptide quantification, targeted proteomics requires the use of stable-labeled peptide standards as internal calibrators or unlabeled peptides as external calibrators. Untargeted or global proteomics allows indiscriminate simultaneous quantification of all detectable proteins (several thousands in a sample) enabled by HRMS and specialized software and databases (Cox and Mann, 2008; Demichev et al., 2020). The software used in global proteomics analysis employs hybrid peptide identification strategies or library-free spectral matching, accurate mass determination, and statistical analysis to identify peptides, quantify proteins, and detect post-translational modifications. HRMS data can be acquired in either data-dependent acquisition (DDA) or data-independent acquisition (DIA) modes. DDA selects and fragments precursor ions based on predefined criteria (typically peak intensity), allowing for the identification of many peptides but may miss low abundance ones. On the other hand, DIA systematically fragments all precursor ions within a defined mass range, offering more comprehensive and reproducible quantitative proteomic data. While DDA is advantageous for broad peptide identification, DIA excels in consistent quantification across multiple runs, making the choice between them dependent on the aim of the research and the balance between proteome coverage, quantification accuracy, and data complexity (Li et al., 2021). In general, targeted proteomics excels in throughput and precision for predefined targets, while global proteomics provides a broader, exploratory quantitative view of the proteome (Prasad et al., 2019; Ahire et al., 2022; Shen et al., 2023). Because of the nature of targeted analysis, parallel reaction monitoring or multiple reaction monitoring proteomics methods can often be more sensitive for low abundance proteins as compared with global proteomics analysis (Heil et al., 2021; Fu et al., 2023). However, HRMS offers a high degree of confidence in analysis of complex samples with high background noise. The pros and cons of such techniques as well as their typical applications within drug development have previously been summarized (Prasad et al., 2019).
Summary of quantitative proteomic techniques (A) and current and future applications in translational pharmacology and precision medicine (B). Abbreviations: EV, extracellular vesicles; MIC, minimum inhibitory concentration; MRM, multiple reaction monitoring; PD, pharmacodynamics; PK, pharmacokinetics; PRM, parallel reaction monitoring.
Recent trends highlight the growing significance of global quantitative proteomics, indicating the equitability of sensitivity and reproducibility between targeted and global proteomic data (Wiśniewski, 2017). The total protein approach (TPA) stands as an emerging strategy to measure protein levels in complex biologic samples utilizing global proteomic data (Wiśniewski and Rakus, 2014). In TPA, protein abundance is calculated based on the assumption that the total MS signal from all proteins reflects the total protein and the total MS signal from a given protein (the sum of the signal from the protein’s proteolytic peptides) corresponds to the partial abundance of the protein in the whole sample (Wiśniewski, 2017). For this, data normalization is crucial for accurate quantification of protein levels across complex biologic samples. TPA employs various normalization techniques such as total intensity normalization and median normalization. Total intensity normalization adjusts for variations in sample loading and instrument sensitivity, while median normalization reduces the impact of outliers by using the median intensity across all samples as a reference. Normalization to housekeeping proteins accounts for variations in sample preparation and loading by comparing the intensities of proteins of interest to stable reference proteins. Additionally, TPA considers experimental design factors to minimize systematic biases and ensure robust normalization of the data, enabling reliable comparisons between samples, batches, and conditions in proteomic studies. Unlike relative quantification methods, the TPA approach aims to quantify the abundance of all proteins present in a sample, facilitating a more comprehensive and precise quantitative understanding of the proteome. This approach circumvents limitations associated with traditional relative quantification, providing critical insights into molecular and cellular processes, biomarker discovery, and disease mechanisms. However, non-unique peptides are used with unique peptides in TPA analysis, and therefore it may be prone to false positive identification and overestimation in quantification (Vasilogianni et al., 2022b). If the protein identification and quantification are based on surrogate peptides unique to individual proteins, global label-free quantification holds immense promise for advancing our understanding of systems pharmacology and mechanistic toxicology of drugs. Invariably, proteomic measurements of the abundance of proteins involved in pharmacokinetics require verification against activity measurements (Achour et al., 2018; Hammer et al., 2020; Rodrigues et al., 2022), especially in the case of novel techniques, new target proteins, or new biologic matrices.
What are the Typical Applications of Proteomics in Translational Pharmacology?
Fig. 2B shows a summary of proteomic applications. In vitro models used in drug metabolism and transport studies (e.g., for in vitro to in vivo extrapolation (IVIVE)) can be validated for protein abundance using quantitative proteomics. The technology is not only useful for protein abundance measurement, but it can also be used to assess quality and batch-to-batch variability of in vitro reagent preparation [e.g., microsomes and membrane preparations, (Xu et al., 2018; Leeder et al., 2022)] by detecting marker proteins and identifying contaminants in sample processing [e.g., placental preparations (Kruger et al., 2023)]. A recent study by Handin et al., demonstrates that a proteome, despite containing less data than the transcriptome, can accurately inform cell type deconvolution using different algorithms (Handin et al., 2023). By analyzing proteomes from cell lines, primary liver cells, and biopsies, accurate deconvolution was achieved, providing insight into extracellular compartments. Applying this to liver biopsies from patients undergoing gastric bypass surgery revealed correlations between immune and stellate cell proportions, inflammatory markers, and early-stage fibrosis markers (Handin et al., 2023).
Similarly, understanding of interspecies differences is critical for interpreting drug toxicology data and allometric scaling of drug pharmacokinetics. Quantitative proteomics has been leveraged to identify interspecies differences in transport and metabolism, such as renal and hepatic transporters and enzymes (Wang et al., 2015; Basit et al., 2019; Sharma et al., 2023). For example, quantitative proteomics data confirmed the absence of BCRP and OCT1 proteins in human kidneys as compared with rodents (Basit et al., 2019). Similarly, unlike humans, significant sex-dependent expression of transporter proteins has been confirmed by quantitative proteomics in rodents (Basit et al., 2019).
In broader terms, many groups have used proteomics for characterization of interindividual variability to investigate the impact of age (Bhatt et al., 2018, 2019; van Groen et al., 2018; Goelen et al., 2023), genotype (Dalton et al., 2020; El-Boraie et al., 2022), and disease conditions (Al-Majdoub et al., 2020; Alrubia et al., 2022; Vasilogianni et al., 2022a). For example, quantitative proteomic analysis of 455 human liver samples revealed that variability in UGT2B17 is associated with age, sex, race, and genotype (Bhatt et al., 2018). Similarly, recent studies have used proteomics for characterizing the effect of diseases or abnormal physiologic conditions, such as Crohn’s disease (Alrubia et al., 2022), colorectal cancer (Vasilogianni et al., 2022a), liver diseases (Prasad et al., 2018; Al-Majdoub et al., 2020; Vasilogianni et al., 2022b), and obesity (Wegler et al., 2022). Not confined to proteomics of the host, the technique extends to quantifying gut microbial proteins, such as glucuronidases, and establishing correlations, such as those between glucuronidase enzymes and the deconjugation rate of drug glucuronides (Parvez et al., 2021).
What are the More Recent Applications of Quantitative Proteomics?
Single-cell proteomics is evolving into a routine tool, facilitating precise analysis of biologic alterations at individual cellular levels, thereby proving instrumental in drug discovery, assessing drug effects on individual cells, enabling precision medicine, and delving into single-cell spatial proteomics, including in understanding drug metabolism (Wheeler et al., 2023). The identification of cellular heterogeneity through proteomic markers contributes significantly to this field (Mund et al., 2022). Beyond small molecule drugs, the technique has shown promise for applications in characterizing variability in proteins involved in the disposition of biologics (Barber et al., 2023). Further, proteomics also holds promise in quantifying proteins at low levels, utilizing biopsy samples or small tissue amounts as well as liquid biopsies. In particular, application of proteomics in quantifying extracellular vesicles isolated from biologic fluids, such as plasma and urine, has a vast potential in precision pharmacotherapy (Rodrigues and Rowland, 2019; Rowland et al., 2019; Achour et al., 2021, 2022).
Lastly, quantitative proteomics allows the quantification of pharmacodynamic markers. For example, the applications of the technique in quantification of receptor tyrosine kinases involved in cancer metastasis (Vasilogianni et al., 2023) and warfarin-induced posttranslational changes (descarboxylation) of prothrombin (Singh et al., 2023) have promising uses in precision medicine. Such proteomic analysis assists in predicting individual patient responses to therapies, minimizing adverse effects, and optimizing treatment outcomes. Integrating proteomic data with other ‘omics’ information offers complementary understanding of an individual's molecular profile, paving the way for personalized therapeutic strategies. These integrated data can be useful in informing quantitative systems pharmacology models, which extend beyond accounting for drug exposure to assess how variability in exposure propagates to changes in response, especially under disease conditions (Musante et al., 2017).
Sample Preparation for Quantitative Proteomics
Different Tissues Have Different Issues
The proteome is quite dynamic, with increasing levels of information and complexity as we move from genes to mRNA to proteins to proteoforms (Bludau and Aebersold, 2020; Smith and Kelleher, 2013). From the ‘one gene encodes one protein’ hypothesis, >20,000 protein-encoding genes are expected, but when considering alternative splicing, alternative proteome promoter usage, and messenger RNA editing, >100,000 transcripts can be produced before post-translational modification (Roth et al., 2005), which can lead to at least a million proteins or proteoforms (Smith and Kelleher, 2013). Quantitatively, the range of protein concentrations in cell lines/tissues is approximately seven orders of magnitude (Beck et al., 2011). The challenge is in detecting and quantifying the low and ultra-low abundance proteins in a reproducible manner across samples (Anderson and Anderson, 2002). The composition of whole proteomes can be obtained by either of two proteomic approaches (top-down and bottom-up). Top-down proteomics that deals with analysis of individually separated intact proteins is cumbersome and offers limited sensitivity and quantitative applications, while bottom-up proteomics has major applications for protein identification and characterization. The complexity of the bottom-up approach (analysis of digested proteins) is a major challenge in proteomic analysis even when using high-resolution peptide separation methods. Modern LC-MS instruments can reproducibly identify at least two-thirds of the proteins expressed in a system, and proteins are identified with a sequence coverage ranging up to 80%. However, this outcome is influenced by various factors, including the type of sample, sample preparation, choice of proteolytic enzymes, fractionation methods, and the biologic matrix (Kim et al., 2014; Yang et al., 2018; Niu et al., 2022). Another limitation with tissue specimens is the post-mortem time, which, if extensive, can significantly affect the content and activity of the proteome (Hansen et al., 2019; Kocsmár et al., 2023). To capture these complexities, efficient and reproducible sample preparation is crucial, especially when dealing with different types of tissues (hard versus soft), various types of cells (rich in proteins or lipids), and different cellular compartments (membrane or cytosolic). There is no one standard approach for preparing protein samples. The protocols differ depending on tissue type, proteome complexity, nature of proteins, and their cellular location. To start a bottom-up proteomic approach, a solubilized protein mixture must be obtained. This covers the following steps: homogenization, protease inhibition, sonication, protein extraction/precipitation, reduction and alkylation, protein digestion, and for low abundance membrane proteins, subcellular fractionation.
Sample Preparation Tools and Protocols
Subcellular sample preparation, which includes both homogenization (cell disruption) and then fractionation, is a critical step in routine proteomic sample preparation (Tastet et al., 2003). Minimum and gentle homogenization should be used to release cell components without degrading protein structures. The choice of homogenizer depends on the tissue type (soft or hard), and tissue homogenization involves shearing, cavitation, or turbulence in a lysis buffer (Huber et al., 2003). For example, a Potter homogenizer is suitable for soft tissues, such as brain and liver (Toni et al., 2019). However, a common method for brain tissue dissociation is enzymatic treatment, which is less efficient compared with Potter homogenizer disruption with, for example, dextran treatment followed by centrifugation (Al-Majdoub et al., 2019). For brain specimens, the fat needs to be removed from the tissue, otherwise mass spectrometry analysis will be sub-optimal. Other homogenizers include mechanical devices (Smith and Xu, 2012), which are suitable for many soft or hard tissue types, e.g., kidney cortex and intestine. An effective way to prepare hard tissue, such as the skin, is bead-based homogenization (Yagi et al., 2020). Cell lysis can also be performed using a pressure cycling homogenizer, but only soft tissues can be effectively lysed with this method; hard tissues remain partially lysed (Cai et al., 2022). Sonication is an important follow-up step to homogenization, which enables cell disruption in suspension (Jin et al., 2021).
During tissue disruption, certain enzymes, such as phosphatases and proteases, are released, and they can alter the structure of proteins. This is especially the case with intestinal fractionation. The intestine is unique in that it requires the use of low temperature and protease inhibitors with (Grangeon et al., 2021) or without phenylmethylsulfonyl fluoride, a known inhibitor of esterase activity (Xie et al., 2002), during tissue processing. This is due to the detrimental effects of the high levels of endogenous proteases present in the lumen and the mucus of the intestine. Without protease inhibitors, proteases can catabolize proteins as enterocytes are homogenized, and this can lead to decreased proteome integrity and protein activities. Additional steps after homogenization include protein depletion or enrichment of selected proteins, but this is not always necessary and is specific to the protein sample [e.g., depletion of plasma albumin before proteomic analysis (Ignjatovic et al., 2019)]. All steps in a protocol ultimately contribute to the quality and reproducibility of protein identification and quantitation as each extra step in sample preparation/fractionation often leads to loss of proteins or changes in proteome composition (Harwood et al., 2014; Wegler et al., 2021). One way to address sample complexity and reduce variability in proteomics is to create standardized, scalable, and parallelized sample preparation protocols, while allowing flexibility to contend with tissue/cellular diversity (Prasad et al., 2019; Varnavides et al., 2022).
Applications in Specific and Disease Populations
Diversity in Patient Populations
The diversity in drug exposure and response in different populations leads to the trend that one drug dose does not fit all. To this end, recent US Food and Drug Administration guidance for the industry suggests considering enrolment of more diverse patient populations in drug development (https://www.fda.gov/regulatory-information/search-fda-guidance-documents/enhancing-diversity-clinical-trial-populations-eligibility-criteria-enrollment-practices-and-trial). Furthermore, workshops and white papers have discussed the importance of specialized drug dosing recommendations and how this can be facilitated by predictive models (Younis et al., 2017; Powell et al., 2021). In such predictive models, drug-dependent parameters (e.g., lipophilicity, permeability, and kinetics), system-dependent parameters (e.g., organ blood flow, organ volume, and protein binding), as well as protein levels of drug transporters and drug metabolizing enzymes in relevant tissues are important for scaling in vitro data to in vivo pharmacokinetics (Sharma et al., 2020). This highlights the importance of elucidating variability in different populations, not only in terms of physiologic parameters, but also in terms of differences in protein levels.
The Areas of Focus so Far
Efforts have been directed to investigating differences in specific populations. To highlight some examples, several renal uptake transporters, such as OAT1, OAT3, OCT2, and URAT1 have been reported to increase in abundance with increasing age from birth (Cheung et al., 2019). Key changes in uptake transporters were observed in the first 2 to 12 years of life, but these also slowly increased to adulthood (above 17 years old). Similarly, renal protein levels of the efflux transporter P-gp (MDR1) were statistically higher after 2 years of age. In contrast, other kidney efflux and uptake transporter levels (e.g., BCRP, MATE1, and GLUT2) were unchanged with age (Cheung et al., 2019). Drug transporters and enzymes also increased with age in the jejunum and ileum. For example, the efflux transporters BCRP and P-gp (MDR1) showed higher abundance in adult jejunum and ileum compared with levels in younger donors (below 18 years old). Intestinal uptake transporters OATP2B1 and OCT1 were, however, unchanged across age groups. Increased protein levels were also observed for CYP3A4 and UGT1A1 in jejunum and ileum with age (Kiss et al., 2021).
Different disease states also influence tissue protein levels. For example, inflammatory bowel disease has been shown to affect levels of intestinal drug transporter and drug metabolizing proteins. Several drug transporters, such as ABCC3 and MCT1, decreased in both inflamed and adjacent non-inflamed colon tissue in ulcerative colitis and Crohn’s disease (Erdmann et al., 2019; Alrubia et al., 2022). A similar decrease was also reported in levels of several CYP enzymes in ileum in Crohn’s disease (Alrubia et al., 2022). Although the studies included very few donors (n < 5), similar results were observed in both cohorts. Furthermore, protein levels of CYP enzymes were markedly decreased in cirrhotic livers from alcoholic and hepatitis C livers (Prasad et al., 2018) and progressively declined with increased severity of cirrhosis (El-Khateeb et al., 2021a,b). Similarly, hepatic transporter levels also decreased with increased severity of liver cirrhosis (Drozdzik et al., 2020; El-Khateeb et al., 2021a). Several drug transporting proteins and drug metabolizing enzymes were also lower in both normal and tumorous liver tissues in donors with colorectal cancer metastasis and chronic hepatitis C with hepatocellular carcinoma, as compared with livers from healthy donors (Billington et al., 2018; Vasilogianni et al., 2022a). Similarly, renal phase II drug metabolizing enzymes (UGT1A9 and UGT2B7) have also been reported to have lower protein levels and activity in tumoral compared with normal kidney tissue (Margaillan et al., 2015).
Small sample size studies indicated potential effects of dementia on brain drug transporters. When comparing transporter levels from the blood brain barrier of donors with Alzheimer's disease (n = 5) and dementia with Lewy bodies (n = 5) with control donors, most of the transporters were unchanged. However, small changes were observed for transporters, such as GLUT1, MCT1, and OAT1, in the blood brain barrier (Al-Majdoub et al., 2019), suggesting that drug transport to and out of the brain mass may not be affected in dementia.
The effect of obesity on drug transporters and drug metabolizing enzymes has been studied with a relatively larger number of donors (n = 38). This revealed small changes in hepatic protein levels of drug transporters and drug metabolizing enzymes in obese compared with lean donors, such as increased OATP1B1 and increased CYP1A2 and CYP2C19 levels in obese donors. Importantly, these changes were smaller than the inter-individual variability of the protein levels (average fold difference of 1.2 between donors with and without obesity as compared with average fold difference of 7.9 across all donors) and were not reflected in changes in in vivo clearance of rosuvastatin (OATP1B1) and caffeine (CYP1A2) (Kvitne et al., 2022; Wegler et al., 2022; Hovd et al., 2023).
Enablers of Prudent Extrapolation to In Vivo Drug Clearance
In addition to quantitative proteomic data, appropriate biologic scalars are required to enable translation from in vitro data measured in various in vitro systems (e.g., microsomes, cytosol, rhCYP and hepatocytes) to in vivo consequences (Neuhoff et al., 2021). Scaling factors are dependent on the type of in vitro system, the species, and tissue source. The generated in vitro parameters must be expressed in units that are scalable for a particular application. For instance, in vitro liver enzyme Vmax expressed in µmol/min/mg of cytosol protein from hepatic cytosolic assays can be translated to in vivo liver enzyme Vmax expressed in µmol/min/kg body weight for usage in a PBPK model. In this IVIVE example, physiologic parameters, such as liver mass and body weight of the specific species are combined with cytosolic protein content per gram liver. This process also applies to scaling factors in kidney and intestine (Harwood et al., 2013; Scotcher et al., 2017).
Commonly used IVIVE scaling factors, such as microsomal, cytosolic, and homogenate protein per gram liver have been shown to change in specific populations. For example, pediatric microsomal protein per gram liver has been reported to increase with age until adulthood (Barter et al., 2008; Leeder et al., 2022), while decreasing in tissues from donors with liver cancer, cirrhosis, steatohepatitis, and inflammation (Billington et al., 2018; El-Khateeb et al., 2020; Vasilogianni et al., 2021; Sierra and Achour, 2024). Similarly, cytosolic protein per gram liver and homogenate protein per gram liver decreased in cancer and inflamed tissues (El-Khateeb et al., 2020; Vasilogianni et al., 2021).
Future Areas of Focus
Further studies are needed to elucidate the diversity in specific populations to fully understand how protein levels are altered and thus affect drug disposition. More information is needed about protein levels in unexplored diseases and in specific populations, such as rheumatoid arthritis, diabetes, pregnancy, elderly, as well as how chronic disease states impact other organs than just the immediately affected organ (e.g., how hepatic drug transporters and metabolizing enzymes are affected by chronic kidney disease). Importantly, studies with larger cohorts are important to also include the inter-individual variability within specific populations in predicted changes in drug exposure.
Individual Patient Characterization for Precision Therapeutics
Catering for Sub-Groups versus an Individual Patient
Between-patient variability in drug exposure and response is affected by the interplay between several internal and external factors, including those that affect the individual patient’s physiology and expression of proteins involved in drug metabolism and disposition, such as age, sex, and disease (Huang and Temple, 2008). Quantitative assessment of pharmacokinetic (PK) variability requires first defining and then characterizing the main parameters affecting drug exposure in the individual as opposed to the average person in a subgroup of patients. In addition to biologic factors, apparent PK variability can also be affected by factors related to the prescriber’s and individual patient’s preferences (Forni Ogna et al., 2017). For characterization of absorption, distribution, metabolism and excretion (ADME) pathways relevant to the drug PK, one or a combination of several methods can be used.
Genotyping can identify a pharmacogenetic category for the patient, loosely linked to a specific activity score (genotype–phenotype correlation), such as CYP2D6 activity brackets (Gaedigk et al., 2008). The Clinical Pharmacogenetics Implementation Consortium publishes guidelines for managing dose adjustment for several drug-pharmacogene pairs, e.g., tacrolimus-CYP3A5 and clopidogrel-CYP2C19, based on genotype information (Scott et al., 2011; Birdwell et al., 2015). However, wide population variations in activity have been reported for each genotype, and some enzymes with large phenotype variability in the population do not have significant genotype diversity that can explain such differences in activity (Klein and Zanger, 2013). Therefore, phenotyping with exogenous probes and endogenous biomarkers is used to better characterize patients for important metabolic and transport pathways. Established probe cocktails include the Geneva cocktail (Bosilkovska et al., 2014), the Cooperstown cocktail (Chainuvati et al., 2003), the Karolinska cocktail (Christensen et al., 2003), and the Pittsburgh cocktail (Frye et al., 1997), which have a limited scope (targeting CYP3A, CYP2C9, CYP2C19, CYP2D6, CYP2B6, CYP2E1, CYP1A2, NAT2, and P-gp) and they overlap in specificity. Endogenous compounds used as markers for individual enzymes and transporters include endogenous metabolites or metabolite-to-parent ratios measured in plasma or urine. Examples include the ratio of 4β-hydroxycholesterol to cholesterol in plasma and the ratio of 6β-hydroxycortisol to cortisol in urine for CYP3A activity, or the use of coproporphyrin I and III as plasma markers for the function of OATP1B1/3 (Mariappan et al., 2017). Although these markers are less invasive than exogenous probes, only a limited number of sufficiently selective endogenous markers have so far been identified (Mariappan et al., 2017; Galetin et al., 2024), and these compounds tend to explain only a fraction of the variability in target protein activity (Diczfalusy et al., 2011).
Liquid Biopsy with Systems Pharmacology Modeling
Systems pharmacology approaches rely on the use of quantitative proteomics to characterize ADME pathways in tissue (either post-mortem, surgical surplus, or tissue biopsy). Whereas tissue proteomics is able to quantitatively assess large numbers of ADME proteins (thousands of targets in one experiment), access to human tissue is precluded by ethical, logistic and legal restrictions. Therefore, recently introduced novel sampling by liquid biopsy offers an alternative that enables acquisition of ‘systems’ data compatible with PBPK models. In broad terms, liquid biopsies can be used for diagnostic, companion diagnostic, or therapeutic applications. Extra-cellular vesicles or exosomes released by tissue into a biologic fluid, such as the blood, contain a time-averaged sample (rather than a snapshot) of the biomolecular composition of their tissue of origin (including DNA, RNA, proteins, lipids, and metabolites) (Achour and Rostami-Hodjegan, 2022). ‘Omics’ analysis generates quantitative data that should reflect tissue levels of the enclosed ADME proteins. In addition to monitoring ADME proteins, liquid biopsy may also be used to define variability in receptors and other drug targets (pharmacodynamic variability) (Rostami-Hodjegan and Achour, 2023). Liquid biopsy studies (Achour et al., 2021, 2022; Barber et al., 2023) demonstrated the possibility of monitoring >500 ADME targets (including 171 enzymes, 362 transporters and FcRn) and >80 drug targets after appropriate correction for individual differences in exosome shedding. Determination of variability in the rate of shedding becomes important when the patient population includes heterogenous cohorts with different diseases or variable severities of the same disease (Jackson et al., 2023).
Applications of Liquid Biopsy in Precision Dosing
With additional validation, wider application of liquid biopsy should facilitate implementation of model-informed precision dosing; the generated quantitative data linked to tissue expression is compatible with modeling platforms, such as Virtual Twins (Polasek and Rostami‐Hodjegan, 2020; Darwich et al., 2021). Virtual Twins are quantitative systems pharmacology models individualized with demographics, genotype data, PK/ pharmacodynamic expression grades (e.g., abundance data from liquid biopsy), and clinical scores (e.g., estimated glomerular filtration rate and hepatic function tests), which go beyond information for the disease cohort to which the individual patient belongs (El‐Khateeb et al., 2021). The use of systems data derived from liquid biopsy with such models offers the possibility of a priori dose selection (initial dose) and dose adjustment (subsequent doses) as well as identification of patients at risk of severe adverse drug effects or therapeutic failure. This application is fully aligned with recent calls for better “patient characterization” in clinical pharmacology drug development and therapeutic practice (Polasek and Peck, 2024). Efforts aimed at verification include work by (Achour et al., 2022), which demonstrated correlation with in vivo activity (of CYP1A2, 2B6, 2C9, 3A, and P-gp) in a cohort with cardiovascular disease phenotyped with the Geneva cocktail, in line with earlier findings by (Rowland et al., 2019) for CYP3A4 in healthy individuals. Applications have so far focused on precision dosing to reduce between-patient variability in drug exposure (Achour et al., 2021; Rostami-Hodjegan et al., 2024) and on assessment of induction drug–drug interaction potential (Rodrigues et al., 2021).
Challenges and Limitations of Liquid Biopsies
Despite its potential, liquid biopsy aimed for therapeutic applications is still in its infancy, with many unknowns and challenges. Recent work has therefore mainly focused on monitoring ADME targets in readily accessible and fairly well-studied systems, such as plasma (Conde-Vancells et al., 2008; Gotanda et al., 2016; Kumar et al., 2017; Rowland et al., 2019; Achour et al., 2021), while the use of more challenging biofluids, such as urine and cerebrospinal fluid, is not common. Selectively collecting extracellular vesicles originating from the liver is a major sample preparation challenge. Published methods tend to either collect all released vesicles, e.g., with resin-based precipitation or ultracentrifugation (high recovery, low selectivity), while applying a mathematical correction for liver shedding and targeting liver-enriched genes (Achour et al., 2021), or enriching liver-derived extracellular vesicles, e.g., with immunoprecipitation (low recovery, high selectivity) (Rodrigues et al., 2021), followed by analysis of RNA and protein content. To date, there is no available method that can achieve high recovery and highly selective extracellular vesicle profiles from specific tissues. More importantly, additional work is required to address the sensitivity challenge (abundance of targets in exosomes is low) and contend with establishing a quantitative link in relation to protein expression of ADME targets in the liver and other tissues. A knowledge gap also remains in the characterization of exosome-derived PK/ pharmacodynamic biomarkers in patients from understudied populations, such as pregnancy and pediatrics, and more importantly, validation efforts in real-world clinical settings are limited.
Conclusions and Outlook
The role of quantitative proteomics in disciplines such as DMPK, PBPK modeling, precision medicine, and translational pharmacology, particularly in quantitative systems pharmacology, is undeniably important and transformative. By providing a comprehensive profile of protein abundance data, post-translational modifications, and protein–protein interactions, quantitative proteomics enables a deeper understanding of disposition, efficacy, and toxicity of drugs at the molecular level as well as interindividual variability and disease effects. This knowledge is instrumental in refining drug development processes, optimizing dosing regimens, and advancing personalized medicine approaches. Moreover, the integration of proteomic data with computational modeling techniques enhances predictive capabilities in pharmacokinetics, pharmacodynamics, and disease progression modeling, thereby facilitating the translation of preclinical findings into clinical applications. Ultimately, quantitative proteomics stands as an indispensable tool in modern pharmacological research, driving innovation and progress toward safer, more effective, and personalized therapeutic interventions (Prasad et al., 2019).
With the renewed interest in application of modeling and simulation by the pharmaceutical industry and increased acceptance of ‘virtual trials’ by regulatory agencies, the number of drug labels informed by modeling and simulation has continued to increase. These cases highlight the increased use of simulations to complement clinical studies or to inform specific applications that would have otherwise been impossible to conduct in real patients. We predict that the integration of proteomics-informed models will continue to improve, and implementation will expand to clinical practice in the near future, especially in the area of personalized therapeutics at the point of care. For such implementation to be realized, specific ‘systems’ data, including alterations over time, with disease progression and during treatment, will be required (Neuhoff et al., 2021). Quantitative proteomics studies should consider the following strategies for improved integration of proteomic data in model-informed clinical and pharmacokinetic applications: (i) selection of the proteomic methods should be determined by the intended application, following ‘target identification-target validation-target quantification’ paradigm, (ii) sufficient validation of proteomic data for precision, reproducibility, linearity, and sensitivity (limit of detection/quantification) is critical; (iii) IVIVE expression-based scalars should consider the same proteomic workflow and a similar proteomic matrix for the in vitro and tissue systems; (iv) quantitative workflows (particularly for sample preparation of different biologic matrices) should be standardized, and such information should be readily shared between centers; (v) quantification of variability should distinguish technical variability from biologic variability (this will require consideration of the required number of individual samples as well as the number of technical and analytical replicates); (vi) continued efforts are required to establish and verify IVIVE-PBPK approaches utilizing activity-protein abundance relationships for key enzymes and transporters in relevant organs to accurately represent in vivo attributes; (vii) dedicated studies to establish IVIVE protein expression-based scalars in unexplored diseases and specific populations, such as diabetes, pregnancy, the elderly, and pediatrics as well as chronic liver and kidney diseases, are required.
Data Availability
This review article contains no datasets generated or analyzed during the current study.
Authorship Contributions
Wrote or contributed to the writing of the manuscript: Prasad, Al-Majdoub, Wegler, Rostami-Hodjegan, Achour.
Footnotes
- Received February 28, 2024.
- Accepted May 22, 2024.
This work received no external funding.
The authors have no actual or perceived conflict of interest in relation to the contents of this article.
Abbreviations
- ADME
- absorption, distribution, metabolism and excretion
- DDA
- data-dependent acquisition
- DIA
- data-independent acquisition
- DMPK
- drug metabolism and pharmacokinetics
- HRMS
- high-resolution mass spectrometry
- IVIVE
- in vitro–in vivo extrapolation
- LC
- liquid chromatography
- MS
- mass spectrometry
- PBPK
- physiologically based pharmacokinetics
- TPA
- total protein approach
- Copyright © 2024 by The American Society for Pharmacology and Experimental Therapeutics