Competition During Catalysis
Abstract
Cytochrome P450 3A4 is known to catalyze the metabolism of both endogenous substrates (such as the 6β-hydroxylation of testosterone) and many important therapeutic agents, including theN-demethylation of erythromycin. However, erythromycin and testosterone have been reported to have little or no effect on the metabolism of each other by recombinant CYP3A4. In an effort to understand the basis of these observations, we studied theN-demethylation of erythromycin and the 6β-hydroxylation of testosterone in human liver microsomes and in microsomes from cells containing recombinant human CYP3A4 and P450 reductase under a variety of experimental conditions. In both human liver microsomal and recombinant CYP3A4 systems, erythromycin inhibited testosterone 6β-hydroxylation in a concentration dependent manner, and vice versa. However, the inhibition mechanism was complex. At low substrate concentrations, testosterone and erythromycin acted as competitive inhibitors to each other. Under these experimental conditions, an apparent competitive inhibition of testosterone 6β-hydroxylation by erythromycin was observed, withKi values similar to that of theKm values for erythromycin. When the rates of testosterone 6β-hydroxylation and erythromycinN-demethylation were determined in microsomal incubations containing both substrates at lower concentrations, the observed rates for each reaction were in good agreement with the calculated rates based on the rate equation describing simultaneous metabolism of two substrates by a single enzyme. However, at high substrate concentrations, the kinetic results could be best explained by a mechanism involving partial competitive inhibition. We conclude from these studies that testosterone and erythromycin mutually inhibit the metabolism of each other, consistent with the fact that CYP 3A4 catalyzes the metabolism of both substrates.
CYP3A41 is one of the major cytochrome P450s in human liver microsomes (1,2). It plays a vital role in the metabolism of both endogenous substrates (such as the 6β-hydroxylation of testosterone) and numerous therapeutic agents, including theN-demethylation of erythromycin and the metabolism of cyclosporin A, nifedipine, and lovastatin. Despite its importance in human drug metabolism, basic knowledge regarding some of the unique characteristics of CYP3A4 is still lacking. Geungerich et al. (3) demonstrated that reconstituted systems containing purified CYP3A4, NADPH-cytochrome P450 reductase, and dilauroylphosphatidylcholine have little or no catalytic activities toward a variety of substrates. To enhance the reaction rates of the CYP3A4-containing reconstituted system, additional components such as cytochrome b5, total lipids, detergent, and glutathione may have to be included, depending on the substrate used (4-9). Several studies suggest that CYP3A4 is an allosteric protein (10,11), although the identity of the allosteric site is unknown. In addition, little is known about the active site topology of CYP3A4. However, it is generally recognized that the active site of this enzyme is spacious enough to accommodate large molecules and even more than one substrate (12).
In recent years, considerable progress has been made towards developing probes to assess the in vivo activity of specific cytochrome P450 isoforms in humans. For example, caffeine has been generally accepted as a specific in vivo probe for CYP1A2 activity (13). On the other hand, a specific in vivo probe for CYP3A has not been totally established or validated. Kinirons et al. (14) have noted the absence of correlation among three putative in vivo probes (erythromycin, dapsone, and cortisol) of CYP3A activity in young healthy men. Similarly, Steinet al. (15) found no correlation between the data from the dapsone urinary recovery ratio assay and the14C-erythromycin breath test in patients with rheumatoid arthritis receiving cyclosporine. It is not clear whether this lack ofin vivo correlation among CYP3A (mainly 3A4) substrates is a result of a unique property of CYP3A4 or because of variable contributions of gastrointestinal metabolism for different probes. Alternatively, CYP3A4 may not be the sole enzyme responsible for the metabolism of these substrates (such as dapsone). To this end, it is interesting to note that in a reconstituted system containing the purified, fused CYP3A4-reductase protein, erythromycin and testosterone (each at 200 μM concentration) did not significantly affect the metabolism of each other (7).
Before one can address the question as to why data from in vivo metabolism for CYP3A4 substrates do not correlate with each other, it is important to make certain that erythromycin and testosterone indeed do not inhibit the metabolism of each otherin vitro. In this paper we describe theN-demethylation of erythromycin and the 6β-hydroxylation of testosterone in human liver microsomes and in microsomes from cells containing recombinant human CYP3A4 and P450 reductase. A wide range of concentrations of each substrate were used to determine whether these two substrates mutually affected the metabolism of each other.
Materials and Methods
Materials.
Testosterone, 6β-hydroxytestosterone, corticosterone, erythromycin, glucose 6-phosphate, NADP, and glucose 6-phosphate dehydrogenase were purchased from Sigma Chemical Co. (St. Louis, MO). All other reagents and solvents were of high analytical grade supplied by Fisher Scientific (Fair Lawn, NJ).
Human liver microsomal preparations kindly provided by Dr. Judy Raucy (Agouron Institute, La Jolla, CA) were used in this study. The organ donors were a 38-yr-old male with no known drug history (HL 24493) and a 34-yr-old male with a history of alcohol and cocaine use (HL 3926). Microsomes were prepared as described elsewhere (16). The pyrophosphate-washed microsomes were resuspended at a protein concentration of 10–15 mg/ml in 10 mM potassium phosphate buffer, pH 7.4 containing 0.25 M sucrose, and frozen at −80°C until used. Protein concentrations and P450 contents were determined using the bicinchoninic acid procedure (17) and according to Omura and Sato (18), respectively. Microsomes from cells containing human CYP3A4/OR were obtained from Gentest Corp. (Woburn, MA).
Enzyme Assays.
Testosterone 6β-hydroxylase activity was determined as described (19, 20). Microsomal samples were incubated with various concentrations of testosterone in 100 mM potassium phosphate buffer (pH 7.4) with 1 mM EDTA, 6 mM MgCl2, and an NADPH-generating system consisting of 10 mM glucose 6-phosphate, 1 mM NADP, and 0.35 units glucose 6-phosphate dehydrogenase in a total volume of 0.5 ml. Reactions were performed at 37°C for 10 min with 0.125 mg (HL 3926) or 0.25 mg (HL 24493) of human liver microsomes and at 37°C for 20 min with 0.25 mg of microsomes prepared from human B-lymphoblast cells. After stopping reactions with 5 ml of CH2Cl2, the samples were spiked with 25 μl of 1 mM corticosterone as internal standard, vortexed, and centrifuged at 3,000 × g for 10 min. The organic layer was removed and evaporated to dryness under nitrogen stream. Samples were dissolved in 0.25 ml of methanol and analyzed by HPLC. Aliquots of 50 μl samples were injected directly onto a Zorbax ODS C18 column (4.6 mm × 250 mm, 5 μm, Sigma-Aldrich, Milwaukee, WI) and eluted with methanol (7.5% tetrahydrofuran):H2O (7.5% tetrahydrofuran) by a linear gradient from 35% to 60% in 35 min at a flow rate of 1 ml/min. Chromatographic peaks were monitored with a UV detector at 254 nm. The retention times for 6β-hydroxytestosterone, corticosterone, and testosterone were 8.9, 17.5, and 25.2 min, respectively.
Erythromycin N-demethylation was determined by the method of Tu and Yang (21) with modifications. Erythromycin was incubated at 37°C for 15 min with 0.5 mg (HL 3926) or 1 mg (HL 24493) of human liver microsomes or for 30 min with 1 mg of microsomes from human B-lymphoblast cells in the presence of a NADPH-generating system as described above. Reactions were quenched with 0.05 ml of 25% ZnSO4 and 0.05 ml of 0.3 N Ba(OH)2. After the samples were vortexed and centrifuged at 14,000 × g for 10 min, 0.35 ml of supernatant was transferred to another tube and then mixed with 0.15 ml of a concentrated Nash reagent (15 g of ammonium acetate and 0.2 ml of acetylacetone in 18 ml of 3% acetic acid). The mixture was incubated at 56°C for 30 min and transferred to a 96-well plate. Samples were analyzed by measuring absorbance at 405 nm with Ceres UV 900 HDi spectrophotometer to determine the formation of formaldehyde (22).
Kinetic studies for testosterone 6β-hydroxylation and erythromycin N-demethylation by microsomes were carried out at testosterone or erythromycin concentrations ranging from 10–1000 μM. Experiments were performed under initial rate conditions. Kinetic parameters were obtained from hyperbolic saturation curves by least squares fit of the data to the Michaelis-Menten equation by nonlinear regression analysis, using K.cat computer software program (BioMetallics, Inc., Princeton, NJ). For inhibition studies, testosterone and erythromycin were incubated simultaneously with microsomes in the presence of a NADPH-generating system.Ki values were determined using the Dixon plot (23).
Results
Kinetic Parameters.
Two human liver microsomal preparations, one high (HL 3926) and one low (HL 24493) in CYP3A4 activity, and microsomes from cells containing recombinant human CYP3A4/OR were used to determine kinetic parameters for testosterone 6β-hydroxylation and erythromycinN-demethylation (table 1). TheKm values ranged from 53 to 128 μM for testosterone and 44 to 78 μM for erythromycin. TheVm values were considerably higher for testosterone 6β-hydroxylation (2 to 20 nmol/min/mg) than for erythromycin N-demethylation (from 0.22 to 2 nmol/min/mg).
Kinetic parameters for testosterone 6β-hydroxylation and erythromycin N-demethylation in microsomal preparations
Mutual Inhibition.
The Km for testosterone and erythromycin are not very different; therefore, one should expect mutual inhibition since both substrates are metabolized by CYP3A4. If this is the case, then the reaction rates of each substrate in the presence of another substrate can be predicted from the following equations (24):
In this study, four concentrations (125, 250, 500, and 1000 μM) of erythromycin were incubated either in the absence or in the presence of 50 and 250 μM of testosterone. After incubation, the rates of metabolite formation were independently measured for each substrate. These rates were also calculated from the equations based on the knownKm and Vm, and the concentration of each substrate used in the incubations. As shown in table 2, the observed experimental rates were generally in good agreement with the calculated rates when 125 and 250 μM of erythromycin were used in the incubations. However, at higher substrate concentrations (500 and 1000 μM erythromycin and 250 μM testosterone), the observed reaction rates were substantially higher than the calculated rates, particularly for testosterone 6β-hydroxylation. These results suggest that at lower substrate concentrations testosterone and erythromycin behave as mutual inhibitors in CYP3A4-catalyzed reactions, whereas at higher substrate concentrations additional factor(s) may contribute to the kinetic mechanism involving both substrates.
Reaction rates of microsomal testosterone 6β-hydroxylation and erythromycin N-demethylation determined in the presence of both substrates
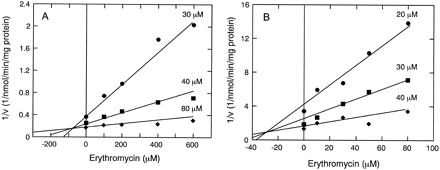
To further explore the nature of interaction between erythromycin and testosterone during catalysis, two types of experiments were conducted to investigate the effect of erythromycin on CYP3A4-catalyzed testosterone 6β-hydroxylation. First, inhibition kinetics of testosterone 6β-hydroxylation by erythromycin were studied at lower concentrations for both substrates (erythromycin, up to 80 μM in one experiment, 600 μM in another experiment; testosterone, up to 40 μM in one experiment, 80 μM in another experiment). Dixon plots indicate that erythromycin competitively inhibited testosterone 6β-hydroxylation with Ki values of 80 μM in HL 3926 (fig. 1A) and 30 μM in CYP3A4/OR (fig. 1B). These Ki values were within the range ofKm for erythromycin determined in separate experiments (see table 1), consistent with the notion that testosterone and erythromycin are alternative competiting substrates for CYP3A4 at these lower substrate concentrations. In the second approach, experiments were designed to determine the effect of erythromycin on the rate of testosterone 6β-hydroxylation; attention was paid particularly at higher concentrations of erythromycin and testosterone. Fig. 2 shows that in both human liver microsomal (figs.2A and 2B) and recombinant CYP3A4 (fig.2C) systems, testosterone 6β-hydroxylation was inhibited by erythromycin in a concentration-dependent manner. At a fixed concentration of 60 μM testosterone, the rate of 6β-hydroxylation could be greatly reduced by high concentrations of erythromycin. However, at higher fixed amounts of testosterone, particularly at 250 and 500 μM concentrations, only partial inhibition of 6β-hydroxylation was observed even at high erythromycin concentrations. These data confirm the results in table 2,i.e., at high concentrations for both substrates, the calculated rates for testosterone 6β-hydroxylation were considerably lower than the observed rates due to partial inhibition by erythromycin.
Dixon Plot for the inhibition of testosterone 6β-hydroxylation by erythromycin.
A. Microsome HL 3926 was incubated with 30, 40, and 80 μM of testosterone and 100, 200, 400, and 600 μM of erythromycin. The estimated Ki value was 80 μM.B. Microsome CYP3A4/OR was incubated with 20, 30, and 40 μM of testosterone and 10, 30, 50, and 80 μM of erythromycin. The estimated Ki value was 30 μM.
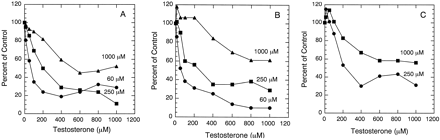
Inhibition of testosterone 6β-hydroxylation by erythromycin in microsomes.
All microsomes were incubated with the indicated testosterone concentrations and various concentrations of erythromycin.A. Microsome HL 3926 was incubated with 60, 125, and 500 μM of testosterone. The corresponding control activities were 3.66, 13.06, and 11.96 nmol/min/mg, respectively. B. Microsome HL 24493 was incubated with 60, 125, and 250 μM of testosterone. The corresponding control activities were 2.79, 4.58, and 4.97 nmol/min/mg, respectively. C. Microsome CYP3A4/OR was incubated with 60 and 250 μM of testosterone. The corresponding control activities were 0.54 and 0.86 nmol/min/mg, respectively.
Detailed inhibition kinetic studies (e.g. determination ofKi for testosterone) for erythromycinN-demethylation were not attempted since the rates of this reaction were low and the colorimetric assay method was not precise enough to measure even lower rates in the presence of testosterone. However, studies were carried out to investigate the effect of testosterone on the rate of erythromycin N-demethylation, including high concentrations of both substrates. Fig. 3shows that in both human liver microsomal (figs. 3A and3B) and recombinant CYP 3A4 (fig. 3C) systems, erythromycin N-demethylation was inhibited by testosterone in a concentration-dependent manner. ErythromycinN-demethylation appeared to be more sensitive to testosterone inhibition even when 1000 μM of erythromycin was used.
Inhibition of erythromycin N-demethylation by testosterone in microsomes.
All microsomes were incubated with the indicated erythromycin concentrations and various concentrations of testosterone.A. Microsome HL 3926 was incubated with 60, 250, and 1000 μM of erythromycin. The corresponding control activities were 0.93, 2.0, and 2.72 nmol/min/mg, respectively. B.Microsome HL 24493 was incubated with 60, 250, and 1000 μM of erythromycin. The corresponding control activities were 0.26, 0.38, and 0.30 nmol/min/mg, respectively. C. Microsome CYP3A4/OR was incubated with 250 and 1000 μM of erythromcyin. The corresponding control activities were 0.28 and 0.34 nmol/min/mg, respectively.
Discussion
Using a reconstituted system containing purified recombinant CYP3A4 and NADPH-cytochrome P450 reductase fusion protein and lipid, Shet et al. (7) recently reported that although both testosterone and erythromycin were metabolized by CYP3A4, they did not significantly inhibit the metabolism of each other when equimolar concentrations (200 μM) of each compound were used in the incubation mixtures. Interestingly, the metabolism of testosterone and erythromycin at the same concentration was strongly inhibited by 200 μM of nifedipine, another substrate of CYP3A4. One would expect mutual inhibition if testosterone and erythromycin were metabolized by the same enzyme. This observation may be explained as follows: (a) It may not always be possible to show the mutual effect of two compounds when substrates are tested at a single concentration. In this respect, experiments conducted at a wide range of concentrations involving both compounds should give more definitive results. (b) Testosterone and erythromycin may have different binding sites on CYP3A4; thus, the presence of one substrate does not significantly affect the metabolism of the other substrate. However, these binding sites cannot be remotely separated on CYP3A4 since both substrates must be close enough to the active oxygen on the heme for oxygenation. Evidence presented by Shouet al. (12) indicates that both phenanthrene and 7, 8-benzoflavone can be present simultaneously in the CYP3A4 active site. (c) The active site topology of the fused protein may not be totally identical to that of the single protein; therefore, the lack of a significant mutual effect may be unique only for the fused CYP3A4.
In an effort to address the question of whether testosterone and erythromycin mutually affect the metabolism of each other by CYP3A4, the metabolism of both compounds was examined under a variety of conditions. Both human liver microsomes and microsomes prepared from human lymphoblastoid cell line containing recombinant CYP3A4 and P450 reductase were used as the enzyme source so that results obtained from one system can be compared with a different system. Our data indicate that in the microsomal preparations used in this study testosterone and erythromycin mutually inhibit the metabolism of each other consistent with the fact that CYP3A4 catalyzes both testosterone 6β-hydroxylation and erythromycin N-demethylation. However, the inhibition mechanism is not purely competitive. Additional mechanisms may contribute to the interaction between these two substrates with CYP3A4, particularly at high concentrations. Although further studies will be required to investigate the nature of testosterone and erythromycin interaction with CYP3A4, we propose the following mechanism, based on partial competitive inhibition (25), to explain our data:
where PT is the metabolite from testosterone (ST); PE the metabolite from erythromycin (SE).
The Km values for testosterone and erythromycin are very similar (50 to 128 μM, see table 1). Therefore, at lower substrate concentrations, e.g. less than 250 μM, testosterone and erythromycin act as competitive inhibitors in the CYP3A4-catalyzed reaction. This conclusion is supported by the following observations. When the rates of testosterone 6β-hydroxylation and erythromycin N-demethylation were determined in microsomal incubations containing both substrates at lower concentrations, the observed rates for each reaction were in close agreement to the calculated rates based on the rate equation describing simultaneous metabolism of two substrates by a single enzyme. Inhibition kinetics of human liver microsomal testosterone 6β-hydroxylation by erythromycin was also analyzed at these concentration ranges. An apparent competitive inhibition was observed with a Ki value of 80 μM. This value corresponded with the Km of erythromycin (78 μM) determined with the same microsomal incubation. AKi of 30 μM for erythromycin was estimated for testosterone 6β-hydroxylation with microsomes containing recombinant CYP3A4, very similar to the Km value of 46 μM for erythromycin N-demethylation determined in the same enzyme system.
At higher substrate concentrations, e.g. greater than 250 μM, testosterone can bind to ESE and erythromycin can bind to EST to form the ESTSEcomplex. This could happen if both testosterone and erythromycin can be present simultaneously in the CYP3A4 active site, and ifKs(TE) and Ks(ET) are substantially higher than Ks(T) andKs(E). Shou et al. (12) have presented evidence that both phenanthrene and 7,8-benzoflavone can simultaneously be at the active site of CYP3A4. Thus, for testosterone 6β-hydroxylation, high concentrations of both substrates will drive the ESTSE formation which can then produce PT and PE. Since k1 is substantially greater than k2, one could assume that k3 is also greater thank4. Therefore, at high concentrations inhibition of testosterone 6β-hydroxylation by erythromycin will be limited. This could explain why in the presence of high substrate concentrations the observed reaction rates for 6β-hydroxylation were substantiallly higher than the calculated rates (table 2) and why high erythromycin concentrations only showed partial inhibition of 6β-hydroxylation at 500 μM testosterone (fig. 2). For erythromycinN-demethylation, greater inhibition was observed by high concentrations of testosterone since ESTSE is favored to produce PT rather than PE.
In summary, evidence is presented that testosterone and erythromycin mutually inhibited the metabolism of each other by CYP3A4 through competitive inhibition at lower substrate concentrations and likely partial competitive inhibition at higher substrate concentrations. Additional experiments will be conducted to test this hypothesis.
Acknowledgments
We thank Dr. J. Raucy (Agouron Institute, La Jollla, CA) for her generous supply of human liver microsomes and Mrs. T. Rafferty for her assistance in preparing the manuscript. We are also grateful to Dr. Charles Y. Huang (National Institutes of Health), Dr. Su Huskey, and Ms. Nancy Thornberry for their valuable suggestions regarding the analysis of kinetic data.
Footnotes
-
Send reprint requests to: Ms. Regina W. Wang, Department of Drug Metabolism, RY 80-A12, Merck Research Laboratories, P.O. BOX 2000, Rahway, NJ 07065.
- Abbreviations used are::
- CYP3A4
- cytochrome P450 3A4
- CYP3A4/OR
- CYP3A4 and NADPH-cytochrome P450 reductase
- Received October 16, 1996.
- Accepted January 20, 1997.
- The American Society for Pharmacology and Experimental Therapeutics