Abstract
The purpose of this work was to evaluate the effect of mutual unbound inhibitor and unbound enzyme depletion on the potency of three antifungal cytochrome P-450 (CYP)3A inhibitors with over 1000-fold range in enzyme affinity. Incubations were performed with human liver microsomal protein concentrations that varied from 25 to 1000 μg/ml. The effect of each inhibitor was evaluated using midazolam as a CYP3A probe. Clotrimazole was found to be a tight binding inhibitor of CYP3A with a Ki of 250 pM. Analysis of percent inhibition data by stepwise linear regression for the matrix of inhibitor and enzyme concentrations used showed that protein concentrations predicted the percent inhibition by clotrimazole (r2 = 0.60, p < .001). When clotrimazole concentrations were added to the model, the r2improved to 0.81, p = .003. Clotrimazole concentrations alone were not a significant predictor of percent inhibition (r2 = 0.21, p = .08). For ketoconazole, protein concentrations provided a weak prediction of the percent inhibition (r2 = 0.39, p = .006). Conversely, ketoconazole concentrations alone were a good predictor of percent inhibition (r2 = 0.55,p < .001). In contrast to results with clotrimazole and ketoconazole, percent inhibition by fluconazole was not dependent on protein concentrations (r2 = 0.06,p = .39). We conclude that microsomal inhibitory potency can be affected by incubation conditions that deplete the unbound concentration of inhibitor available to the enzyme. This may introduce serious error into a quantitative prediction of an in vivo drug-drug interaction based on an in vitro derivedKi value.
The use of steady-state experimental designs in the determination of catalytic and inhibitory constants for cytochrome P-450 (CYP)1 enzymes is widespread despite the fact that a variety of conditions may exist that violate one or more key assumptions that form the basis of this type of analysis. For instance, complications arise when the concentrations of substrate and/or inhibitor in solution are significantly reduced by time-independent factors such as nonspecific binding to other components of the incubation mixture (Obach, 1996) or by mutual depletion (Morrison, 1969; Henderson, 1973; Szedlacesek and Duggleby, 1995). Time-dependent factors such as metabolic depletion, enzyme inactivation (Guengerich, 1997; Voorman et al., 1998), and generation of inhibitory metabolites (Babany et al., 1988; He et al., 1995; Sutton et al., 1997) may also occasionally be of concern. Unfortunately, many of these complexities have only recently been exposed as significant problems in CYP research, despite the fact that the underlying issues have been well recognized in the general literature on both practical and theoretical levels. In this paper we examine the practical implications of a specialized issue associated with inhibitor depletion, that is, depletion of the inhibitor by high-affinity binding to the target CYP enzyme itself, or mutual depletion.
Experimental Procedures
Materials.
Clotrimazole, imidazole, and tritylchloride were purchased from Sigma Chemicals (St. Louis, MO). Methanol and ethyl acetate were obtained from Fisher Scientific Co. (Fairlawn, NJ). Ammonium acetate was obtained from J.T. Baxter (Phillipsburg, NJ). Fluconazole was a gift from Pfizer Inc. (Groton, CT). Ketoconazole was purchased from Research Diagnostics (Flanders, NJ). Midazolam (MDZ), 1′-hydroxymidazolam and 1′-[2H2]-hydroxymidazolam were gifts from Roche Laboratories (Nutley, NJ).
Tissue Collection.
Human liver tissue was obtained from organ donors through the Solid Organ Transplant Program at the University of Washington Medical Center and the Northwest Organ Procurement Agency (Seattle, WA). Liver microsomes were prepared as described elsewhere and stored at −80°C (Paine et al., 1997). Protein concentrations were determined by the method of Lowry et al. (1951). Details regarding donor history have been presented elsewhere (Paine, 1997).
Inhibition Studies.
Exploratory studies with clotrimazole were performed in triplicate for three different human liver microsomal preparations (HL-127, 131, and 141) using 16 μg of protein per 1 ml incubation volume. Clotrimazole (final concentration 1.5 nM) was dissolved in acetone, added to the incubation tube and the acetone was allowed to evaporate. Microsomes, potassium phosphate buffer (pH 7.4), and MDZ (4 μM) were then added. Samples were preincubated at 37°C for 5 min. MDZ 1′-hydroxylation was initiated by the addition of 1 mM NADPH. After 4 min of incubation, the reaction was stopped by the addition of 0.1 M Na2CO3, pH 11. The internal standard 1′-[2H2]-hydroxymidazolam was added to the mixture. Samples were prepared for gas chromatography-mass spectrometry analysis as described previously (Thummel et al., 1994).
To test for depletion of unbound inhibitor by high-affinity binding to microsomal proteins, clotrimazole (1.5, 3, and 6 nM) or ketoconazole (10, 50, and 100 nM) was dissolved in acetone and added to 15-ml reaction tubes. The solvent was allowed to evaporate. Fluconazole (10, 30, and 60 μM) was dissolved in phosphate buffer (pH 7.4) and placed in the reaction tubes. Subsequently, human liver (HL-151) microsomal protein, 25, 50, 100, 200, 500, or 1000 μg, phosphate buffer (pH 7.4), and MDZ (4 μM) were added to a final volume of 1 ml. MDZ 1′-hydroxylation formation velocities were then determined as described above.
To test for metabolic depletion of clotrimazole, the inhibitor (1.5 nM), MDZ (4 μM), phosphate buffer, and microsomes (500 μg) were added to four reaction tubes and preincubated for 5 min. Reactions were initiated by the addition of 1 mM NADPH. Duplicate samples were incubated for 1 and 4 min, respectively. Reactions were terminated by the addition of Na2CO3, pH 11. Samples were prepared for liquid chromatography-mass spectrometry (LC-MS) analysis as follows. The internal standard, tritylimidazole was synthesized by the method of Davis et al. (1982). After the addition of tritylimidazole, samples were extracted twice with 5 ml ethyl acetate. The organic layer was evaporated to dryness and analytes were reconstituted in 150 μl of mobile phase (70:30 methanol:10 mM ammonium acetate buffer, pH 6.8).
Clotrimazole LC-MS Assay.
Liquid chromatography was performed on a Shimadzu LC-10AD solvent delivery system (Shimadzu Scientific Instruments, Inc., Columbia, MD) fitted with a Zorbax Eclipse XDB-C8, 2.1 × 50 mm, 5 μm column (Mac Mod Analytical, Chadds Ford, PA) and a 0.2 mm ODS-C18 Haiguard Guard Disc (Higgins Analytical, Chadds Ford, PA). Analysis was carried out under isocratic conditions (70:30 methanol:10 mM ammonium acetate buffer, pH 6.8) at a flow rate of 0.2 ml/min using a sample injection volume of 40 μl. Interfaced to a Micromass Quatro II tandem quadrupole mass spectrometer (Micromass Ltd., Manchester, UK) via a splitter, 50 μl/min of the effluent was subjected to positive electrospray (+ESP) ionization at a source temperature of 100°C, using nitrogen as the nebulizing and bath gas. Cone and Electrospray probe voltages were 35 V and 3.8 kV, respectively. Selected-ion monitoring data acquisition was accomplished using PC-based Micromass MassLynx 2.1 software (Micromass Ltd., Manchester, UK). Ions monitored were m/z 243.2 and 277.2, corresponding to the neutral loss of imidazole from the [M + H]+ ion of the tritylimidazole internal standard and clotrimazole analyte, respectively, at dwell times of 500 ms per ion. The elution times for tritylimidazole and clotrimazole were 4.3 and 4.5 min, respectively. The limit of detection was 0.5 nM.
Statistical Analysis of Inhibitor Depletion.
The effect of increasing protein concentration on the extent of inhibition of MDZ 1′-hydroxylation with clotrimazole, ketoconazole, or fluconazole coincubation was evaluated both graphically and by regression analysis. For each liver microsomal protein concentration in the incubate, the percent inhibition [(1 − vi/vo) × 100] was graphed as a function of the total nominal inhibitor concentration. In the absence of depletion of unbound inhibitor, the family of curves obtained for each inhibitor should be superimposable. The null hypothesis assumes that they are not. This hypothesis was tested by multiple regression analysis using SPSS version 7.5 (SPSS Inc., Chicago, IL). Because the individual effect of inhibitor and protein concentration are not linearly related to the percentage of inhibition, both predictor (inhibitor and protein concentrations) and response (percent inhibition) variables were log-transformed. Residuals were examined visually to ensure a random distribution. Regression coefficients were considered to be significant when p< .05.
Determination of Clotrimazole Ki.
For a reversible inhibitor, the inhibition constant (Ki) can be expressed as the following:
Results and Discussion
Clotrimazole was found to be a very potent inhibitor of CYP3A4 activity in microsomes prepared from three human livers. When incubations were performed with 16 μg of human liver microsomal protein (1–3 pmol CYP3A4, as determined by Western blot analysis;Gibbs et al., 1999; Paine, 1997) and 3 nM inhibitor, greater than 90% inhibition of CYP3A activity was observed. The estimated ratio of inhibitor to enzyme for individual liver microsomal preparations varied from 2:1 to 0.67:1. This combination of results indicated that a substantial fraction of the clotrimazole present in these incubations must have been bound to the enzyme. By difference, the amounts of clotrimazole that were either free in solution or bound nonspecifically to protein must have been small to negligible. This condition is classically defined as mutual depletion.
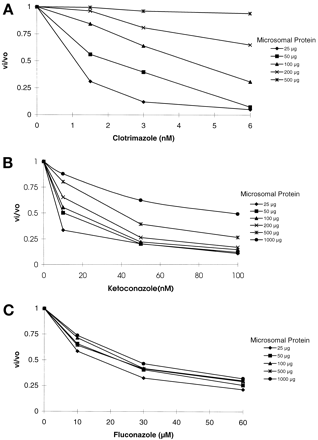
To further investigate this phenomenon, the effect of CYP3A and microsomal protein concentration on the inhibitory effect (vi/vo) of three CYP3A inhibitors (clotrimazole, ketoconazole, and fluconazole) of varying potencies was determined (Fig. 1). We found that, in the absence of inhibitor, product formation was linear with protein concentration up to 200 μg/ml. Because some metabolic depletion of substrate (>6%) occurred at protein concentrations above this level, the inhibitory effects observed at 500 and 1000 μg protein may underestimate the true inhibitory effect. The initial rate (0–4 min) of product formation in the absence of inhibitor declined nonlinearly with increasing protein concentration: 100%, 104%, 101%, 94%, 83%, and 59% (mean percent for three determinations with 25 μg set at 100%) for 25, 50, 100, 200, 500, and 1000 μg/ml protein, respectively. This finding was consistent with the sequestration or metabolism of an appreciable fraction of the unbound MDZ at the highest protein concentrations.
Inhibition of MDZ 1′-hydroxylation at various protein concentrations.
A, clotrimazole; B, ketoconazole; C, fluconazole. For each inhibitor, vi = inhibited velocity; va = uninhibited velocity. Individual data points represent the mean value from duplicate incubations.
As expected, the percent inhibition of MDZ 1′-hydroxylation was found to increase with increasing concentrations of inhibitor. However, the extent of inhibition (1 − vi/vo) was substantially decreased by increasing amounts of microsomal protein for clotrimazole and ketoconazole, but not for fluconazole. For example, as the amount of protein in the incubate increased from 25 to 500 μg, the degree of inhibition with 3 nM clotrimazole decreased from 95% to 6% (Fig. 1A). In the case of ketoconazole, the percent inhibition decreased from 80% to 38% at 50 nM ketoconazole as protein concentrations increased from 25 to 1000 μg/ml (Fig. 1B). In contrast, inhibition of CYP3A by fluconazole was not altered appreciably by protein concentrations up to 1000 μg/ml (Fig. 1C). For fluconazole, the data at high protein concentrations indicate that the masking effects of substrate depletion via metabolic conversion to product were minor when inhibition was less than 60%.
The effect of protein concentration on the inhibition kinetics of the three antifungal agents was analyzed by stepwise linear regression of the log-normalized dependent (percent inhibition) and independent (inhibitor and protein concentrations) variables (Table1). It should be emphasized that the relative weighting of effects of protein and inhibitor concentrations on the partial correlations presented in Table 1 was clearly dependent on the range of the dependent variables and that the range of values used in this experiment were chosen to maximize the effect of protein concentration. Within the matrix of clotrimazole and protein concentrations chosen, protein concentration correlated with the percent inhibition, r2 = 0.60, p< .001. When clotrimazole concentrations were added to the model, r2 improved to 0.81, p = .003. Clotrimazole concentrations alone were not a significant predictor of percent inhibition (r2 = 0.21, p= .08). For ketoconazole, protein concentrations weakly predicted percent inhibition (r2 = 0.39, p= .006). When ketoconazole concentrations were added to the model, r2 improved to 0.94, (p< .001). Conversely, ketoconazole concentrations alone were a good predictor of percent inhibition (r2 = 0.55,p < .001). In contrast to ketoconazole and clotrimazole, analysis by stepwise linear regression showed that the percent inhibition observed with fluconazole was not dependent on protein concentration (r2 = 0.06,p = .39). Fluconazole concentrations alone were an excellent predictor of the percent inhibition (r2= 0.90, p = .001).
Statistical analysis of CYP3A inhibition at various protein concentrations by antifungal inhibitors
The pronounced inverse relationship between inhibitory effect and protein concentration for clotrimazole and ketoconazole is consistent with significant reduction in the concentration of unbound inhibitor. This could be the result of both specific and nonspecific binding at high microsomal protein concentrations and primarily specific binding at low protein concentrations. The possibility that mutual depletion (depletion of both unbound inhibitor and unbound enzyme) played an important role in the generalized protein effect was assessed with the following assumptions: 1) a one to one complex of inhibitor to enzyme is completely inhibitory and 2) additional high-affinity binding sites for the inhibitor are not present in liver microsomes (i.e., all of the inhibitor is either bound to the enzyme or free in solution). Direct LC-MS analysis of the incubates revealed no significant loss of clotrimazole during the 4-min incubation with 0.5 mg microsomal protein and that measured total concentrations (protein bound and unbound) of the inhibitor corresponded to the nominal concentration (1.7 versus 1.5 nM), indicating that all of the inhibitor was intact and potentially available to the enzyme. The fate of ketoconazole not bound to the enzyme is less certain because we did not determine whether it was depleted by metabolism. However, it would appear that metabolism was probably minor because under the most severe incubation condition (1 mg protein) as much as 66% of the inhibitor in a 10 nM dose must have been bound to the enzyme to produce the degree of inhibition observed (Fig. 1B). Also, the incubation interval was relatively short (4 min), limiting ketoconazole metabolism.
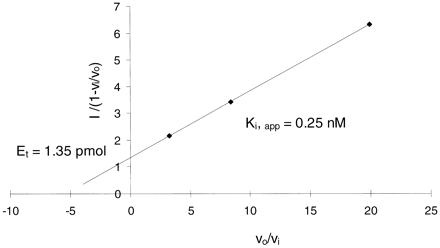
Using eq. 2, which assumes noncompetitive inhibition, theKi for the inhibition of CYP3A4 by clotrimazole was found to be 0.25 nM; the estimate of CYP3A4 content in HL-151 microsomes provided by this transformation was 0.054 pmol/μg protein (Fig. 2). If clotrimazole is a competitive inhibitor, the Ki would be 0.12 nM and enzyme content would be the same (Segel, 1975). We previously measured total CYP3A4 content for HL-151 (apoenzyme and active enzyme) by Western blot analysis and obtained a value of 0.096 pmol/μg protein (Paine, 1997). By inspection of eq. 2, it is clear that the actual enzyme content will be equal to or greater than the graphical estimates, depending on the extent to which inhibitor is not available to enzyme due to sequestration at other microsomal binding sites. Thus, we conclude that the true active enzyme content in HL-151 was somewhere between 0.054 and 0.096 pmol/μg protein.
Determination of clotrimazole Ki.
Individual data points represent the mean result from duplicate incubations. The Ki was estimated based on eq. 2 and the re-plot method of Henderson (1973).Ki,app was generated from incubations with the lowest amount of enzyme and inhibitor reported in Fig. 1. Inhibitor (It) concentrations were 1.5, 3, and 6 nM, and MDZ concentration was 4 μM.
Under any given set of incubation conditions, the amount of inhibitor that is bound to CYP3A4 can be calculated as the product of (1 − vi/vo) and enzyme content in the incubate. For this approach it was assumed that catalytic activity of the enzyme was not observed when inhibitor was bound to the enzyme and that only one high-affinity binding site on the enzyme contributes to removal of free inhibitor in solution. Estimates of the fraction of the total clotrimazole, ketoconazole, or fluconazole pool bound to CYP3A4 [(IE/It) × 100] are presented in Table 2. Using the higher estimate of microsomal CYP3A4 content, the predicted fraction of total clotrimazole remaining in solution when the amount of enzyme was in excess of inhibitor was very small to nil. Using the lower estimate of CYP3A4 content, the predicted fraction of total clotrimazole bound to the enzyme was also considerable (>50%) at all protein concentrations. In the case of ketoconazole, positive evidence for mutual depletion of inhibitor and enzyme was also demonstrated at higher concentrations of protein (Table 2), however, it appeared to be minimal at low concentrations of enzyme. This behavior would be expected in the absence of nonspecific binding, due to the higherKi of ketoconazole, when compared to clotrimazole and the lower amount of enzyme used (see eq. 1). There was no suggestion of unbound fluconazole depletion at all protein concentrations used.
Predicted percent of inhibitor bound to enzyme at various inhibitor and enzyme concentrations
The problem of inhibitor depletion in the characterization of CYP3A4 inhibition kinetics by ketoconazole may be widespread. PublishedKi values for the inhibition of human liver microsomal CYP3A by ketoconazole vary considerably from 4 to 8000 nM (Wrighton and Ring, 1994; Lampen et al., 1995; Bourrie et al., 1996;von Moltke et al., 1996; Gibbs et al., 1999). Our results show that the apparent microsomal inhibitory potency may be affected by lab-to-lab variations in in vitro incubation conditions that can deplete the free concentration of inhibitor available to the enzyme. The most obvious approach to be taken to avoid the problem of inhibitor depletion with high-affinity molecules is to reduce the concentration of enzyme below that of the lowest inhibitor concentration tested. Under this condition, the Ki for ketoconazole and fluconazole were found to be 15 nM and 11 μM, respectively (Gibbs et al., 1999). However, this may not always be achievable because of substrate/product assay limitations. In those circumstances, the approach described for clotrimazole, with independent confirmation of the enzyme concentration, can be applied.
Footnotes
-
Send reprint requests to: Dr. Kenneth E. Thummel, Ph.D., Department of Pharmaceutics, Box 357610, University of Washington, Seattle, WA 98195. E-mail: thummel{at}u.washington.edu
-
This study was supported in part by the National Institutes of Health Grants P01 GM32165, P30 ES07033 (K.E.T. and K.L.K.), and Training Grant GM 07750 (M.A.G.).
- Abbreviations used are::
- CYP
- cytochrome P-450
- MDZ
- midazolam
- LC-MS
- liquid chromatography-mass spectrometry
- Received October 6, 1998.
- Accepted January 19, 1999.
- The American Society for Pharmacology and Experimental Therapeutics