Abstract
Oseltamivir phosphate is an ethyl ester prodrug widely used in the treatment and prevention of both Influenzavirus A and B infections. The conversion of oseltamivir to its active metabolite oseltamivir carboxylate is dependent on ester hydrolysis mediated by carboxylesterase 1 (CES1). We recently identified two functional CES1 variants p.Gly143Glu and p.Asp260fs in a research subject who displayed significant impairment in his ability to metabolize the selective CES1 substrate, methylphenidate. In vitro functional studies demonstrated that the presence of either of the two mutations can result in severe reductions in the catalytic efficiency of CES1 toward methylphenidate, which is required for hydrolysis and pharmacological deactivation. The aim of the present study was to investigate the function of these mutations on activating (hydrolyzing) oseltamivir to oseltamivir carboxylate using the cell lines expressing wild type (WT) and each mutant CES1. In vitro incubation studies demonstrated that the S9 fractions prepared from the cells transfected with WT CES1 and human liver tissues rapidly convert oseltamivir to oseltamivir carboxylate. However, the catalytic activity of the mutant hydrolases was dramatically hindered. The Vmax value of p.Gly143Glu was approximately 25% of that of WT enzyme, whereas the catalytic activity of p.Asp260fs was negligible. These results suggest that the therapeutic efficacy of oseltamivir could be compromised in treated patients expressing either functional CES1 mutation. Furthermore, the potential for increased adverse effects or toxicity as a result of exposure to high concentrations of the nonhydrolyzed prodrug should be considered.
Oseltamivir phosphate (Tamiflu; Roche, Nutley, NJ) is widely used in the treatment and prophylaxis of both Influenzavirus A and B infections. In addition, oseltamivir may be effective in preventing or treating avian influenza or so-called “bird flu.” Oseltamivir is an ester prodrug and, in general, it is readily converted to its active form oseltamivir carboxylate mediated by hepatic carboxylesterase 1 (CES1) (Fig. 1) (Shi et al., 2006). The active metabolite exerts its antiviral effects via the selective inhibition of neuraminidase.
Carboxylesterases are members of the αβ hydrolase -fold family and expressed in many tissues, especially in the liver, small intestine, and lung (Satoh and Hosokawa, 2006; Ross and Crow, 2007). The major human carboxylesterases include CES1 (UniProtKB/Swiss-Prot P23141) and carboxylesterase 2 (CES2) (UniProtKB/Swiss-Prot O00748). CES1 and CES2 are largely distinguished from one another by their substrate specificity and tissue distribution (Imai et al., 2006; Satoh and Hosokawa, 2006). CES1 more readily catalyzes substrates with a relatively large acyl group and small alcohol group such as methylphenidate, temocapril, and oseltamivir (Sun et al., 2004; Imai et al., 2005; Shi et al., 2006). In contrast, CES2 preferentially hydrolyzes compounds bearing a small acyl moiety and bulky alcohol group, which includes agents such as cocaine and irinotecan. CES1 predominates in the human liver, whereas CES2 is the major carboxylesterase expressed in the intestine (Imai et al., 2006). Hepatic CES1 is the major esterase governing the metabolism of numerous and structurally diverse therapeutic agents formulated as carboxylic acid esters, carbamates, thioesters, and amide compounds including those prodrugs formulated as esters. In addition, a number of endogenous substrates are recognized.
In a recent study, we identified two CES1 mutations, p.Gly143Glu and p.Asp260fs (Zhu et al., 2008), in a subject who displayed profound alteration of the pharmacokinetics of racemic (dl)-methylphenidate (Ritalin; Novartis Pharmaceuticals, Summit, NJ), a selective CES1 substrate, during a single-dose pharmacokinetic study (Patrick et al., 2007). The minor allele frequency of p.Gly143Glu was estimated to be 3.7, 4.3, 2.0, and 0% in white, black, Hispanic, and Asian populations, respectively, by a genotyping study that contains a total of 925 subjects with varied racial and ethnic backgrounds. It was concluded that the p.Asp260fs variant was extremely rare because none of the 925 screened subjects carried this mutation. The functional consequences of both mutations were investigated using cell lines stably expressing each individual mutant. The in vitro incubation study demonstrated that the catalytic function of both p.Gly143Glu and p.Asp260fs is impaired to such a significant degree that CES1-mediated methylphenidate hydrolysis was essentially nil using these two CES1 mutants, whereas wild-type (WT) CES1 readily cleaved the ester (Zhu et al., 2008).
Even though the two newly discovered CES1 mutations were determined to be dysfunctional enzymes in terms of hydrolyzing methylphenidate to its inactive metabolite ritalinic acid, the influence of these CES1 variants on prodrug activation has not been examined to date. Oseltamivir (a drug in wide therapeutic use) has recently been shown to be a selective substrate of CES1 (Shi et al., 2006), making it an excellent candidate compound to assess the effect(s) of the identified CES1 mutations on prodrug activation. In addition, accumulating evidence has indicated that the biotransformation of oseltamivir phosphate to the primary active form oseltamivir carboxylate is not only related to its antiviral efficacy but also associated with potential toxicity (http://www.fda.gov/cder/foi/nda/2000/21-246_Tamiflu_Pharmr.pdf).
CES1-mediated activation of oseltamivir phosphate.
In the present study, we investigated the influence of the two newly identified CES1 variants on the metabolism (i.e., activation) of the prodrug oseltamivir phosphate. The results suggested that genetic variants of CES1 that result in dysfunctional enzyme activity could likewise play an important role in both therapeutic efficacy as well as tolerability or toxicity during oseltamivir therapy.
Materials and Methods
Materials. Oseltamivir phosphate and its active metabolite oseltamivir carboxylate were obtained from Toronto Research Chemicals Inc. (North York, ON. Canada). p-nitrophenyl acetate (PNPA) and p-nitrophenol (PNP) were purchased from Sigma-Aldrich (St. Louis, MO). All other chemicals and reagents were of the highest analytical grade and were commercially available.
Enzymatic Study. The establishment of Flp-In-293 cells (Invitrogen, Carlsbad, CA) stably expressing WT and p.Gly143Glu and p.Asp260fs CES1 has been described previously (Zhu et al., 2008). The transfected cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and 100 μg/ml hygromycin B. After reaching approximately 95% confluence, cells were then washed and harvested in reaction buffer (phosphate-buffered saline containing 10 mM HEPES, pH 7.4). Afterward, cells were sonicated and then centrifuged at 9000g for 30 min at 4°C. The supernatant (S9 fraction) was collected and stored at –70°C until use. The liver tissues were obtained from a healthy liver donor and determined to express neither p.Gly143Glu nor the p.Asp260fs mutation and served as a native CES1 control. The liver samples (∼300 mg) were homogenized, and the S9 fraction was obtained after centrifugation at 9000g for 30 min at 4°C. The protein concentrations were determined using a Pierce BCA assay kit (Pierce, Rockford, IL).
The oseltamivir hydrolysis study was carried out in 1.5-ml tubes at a total volume of 100 μl. Before incubations, oseltamivir phosphate solutions were freshly prepared in 50 μl of reaction buffer. The reaction was initiated by mixing oseltamivir phosphate with 50 μl of S9 fractions. The final oseltamivir phosphate concentrations ranged from 10 to 5000 μM. Our preliminary study indicated that the formation of oseltamivir carboxylate was linear with a series of S9 protein concentrations (0.05–0.5 mg/ml) and incubation times (5–15 min) that we tested. In the present study, the enzymatic reactions were performed with the final S9 protein concentration standardized at 0.1 mg/ml and an incubation period of 10 min at 37°C. After incubation, the reaction was terminated by adding 500 μl of methanol containing 40 μM ritalinic acid as the internal standard. The mixture was centrifuged at 16,000g for 5 min to precipitate protein, and the supernatants were then analyzed using an established high-performance liquid chromatography (HPLC) assay. Enzyme kinetic data of oseltamivir hydrolysis were fit to the Michaelis-Menten equation, and kinetic parameters Km and Vmax were calculated using nonlinear regression analysis with GraphPad Prism software (GraphPad Software Inc., San Diego, CA). In addition, PNPA, a widely used esterase substrate (including CES1), was included in the study as a positive control using a method described previously (Zhu et al., 2008).
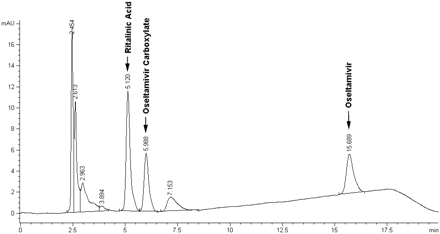
HPLC Analysis. An HPLC method was used to measure oseltamivir carboxylate formation as a consequence of oseltamivir phosphate hydrolysis. The HPLC system consisted of an Agilent 1100 HPLC system (Agilent Technologies, Santa Clara, CA) equipped with a diode-array detector with the wavelength set at 220 nm. The mobile phase was a mixture of methanol and 20 mM KH2PO4 (pH2.5). A gradient elution was applied for the separation with the time program set as follows: from 0 to 4 min, methanol was 44%, and increased to 50% from 4 to 14 min, then maintained at 50% until 16 min, where methanol was returned to the initial condition (44%). Ritalinic acid, oseltamivir carboxylate, and oseltamivir were eluted at 5.1, 6.0, and 15.7 min, respectively, with the flow rate set at 1 ml/min. In Fig. 2, a typical chromatogram is represented of 100 μM of oseltamivir hydrolyzed by WT CES1 S9 fractions after incubation. The intraday and interday relative standard deviations were determined to be less than 10%. The lower limit of quantification of oseltamivir carboxylate was 0.25 μM.
Results
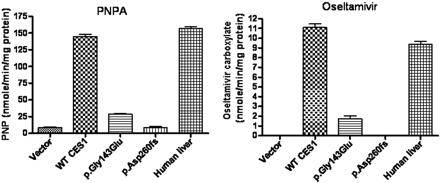
PNPA is a sensitive and established model substrate of CES1 as well as other human esterases. The PNPA hydrolysis assay demonstrated that WT CES1 prepared from the cells transfected with WT CES1 gene rapidly hydrolyzed PNPA to PNP with a catalytic efficiency comparable with that of normal human liver tissues (Fig. 3). Consistent with our previous observations, the enzymatic activity of both p.Gly143Glu and p.Asp260fs toward PNPA was dramatically reduced relative to WT enzyme (Zhu et al., 2008).
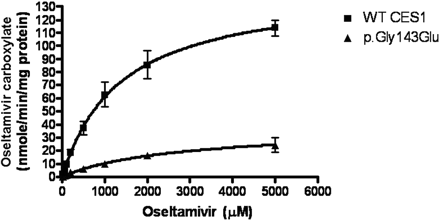
The oseltamivir phosphate incubation study demonstrated that the S9 preparations of both WT CES1-transfected cells and human liver tissues efficiently convert oseltamivir to its active antiviral component, oseltamivir carboxylate, suggesting that oseltamivir serves as an excellent substrate of CES1 (Fig. 3). The Vmax and Km values were determined to be 145 ± 5 nmol/min/mg protein and 1.38 ± 0.13 mM, respectively, under our experimental conditions (Fig. 4). The CES1 variants p.Gly143Glu and p.Asp260fs displayed poor catalytic activity toward oseltamivir hydrolysis (Figs. 3 and 4). The Vmax value of p.Gly143Glu was found to be 37 ± 1 nmol/min/mg protein, which is approximately 25% of that of WT CES1. The Km value of p.Gly143Glu was estimated to be 2.15 ± 0.18 mM. In addition, p.Asp260fs failed to produce any detectable hydrolysis of oseltamivir as measured by the formation of oseltamivir carboxylate (Fig. 3). The human liver S9 fractions prepared from a healthy donor specimen produced similar catalytic activity toward both PNPA and oseltamivir phosphate, which was in excellent agreement with that of our WT CES1-transfected cells (Fig. 3).
Discussion
CES1 is the predominant hydrolase in the liver and plays an important role in the biotransformation of drugs and prodrugs that contains ester bonds. CES1 genetic variants and their potential for having therapeutic implications have been increasingly reported recently. Our previous study identified two nonsynonymous coding region variants, p.Gly143Glu and p.Asp260fs. In vitro functional studies have shown that the catalytic function mediating the typically efficient and rapid hydrolysis of methylphenidate was clearly disrupted in both the p.Gly143Glu variant and the p.Asp260fs mutation. The potential for clinically significant outcomes in the presence of these two mutations was investigated in the original subject found to carry both CES1 variants. That subject displayed an extremely abnormal pharmacokinetic profile after the administration of methylphenidate, displaying vastly higher overall blood concentrations of methylphenidate and an unprecedented distortion in the disposition of the respective isomers of the drug (Patrick et al., 2007; Zhu et al., 2008). In addition, the subject experienced significantly higher cardiovascular vital signs relative to other 19 study subjects serving as a pharmacodynamic correlate to the pharmacokinetic observations (Zhu et al., 2008).
Representative chromatograph of oseltamivir carboxylate. Oseltamivir carboxylate was analyzed by the HPLC assay after incubation of oseltamivir phosphate (100 μM) and WT CES1 S9 fractions at 37°C for 10 min.
Hydrolysis of PNPA and oseltamivir by human liver microsomal, WT CES1, and its mutants p.Gly143Glu and p.Asp260fs. The hydrolytic products of PNPA and oseltamivir were determined after incubating the substrates with the enzymes at 37°C for 10 min. Data were expressed as the mean ± S.D. (n = 4).
Because the activation of many ester prodrugs depends to a great degree upon functional CES1 enzyme to produce the therapeutic moiety, dysfunctional CES1 variants could hinder prodrug activation and lead to the alteration of therapeutic effects and accumulation of the parent prodrug with continued dosing. Such an outcome could lead to therapeutic failure and, depending on the compound administered, unanticipated adverse effects or toxicities. As a prodrug, oseltamivir does not exhibit activity toward the influenza virus unless it is converted to its active metabolite oseltamivir carboxylate by CES1 (Fig. 1). In the present study, the catalytic activity of p.Gly143Glu and p.Asp260fs toward oseltamivir hydrolysis (i.e., activation) was investigated using transfected cell lines stably expressing WT and individual mutant CES1 enzyme. The data indicated that the enzymatic activity of p.Gly143Glu is substantially decreased with a Vmax value approximately one fourth that of WT CES1, whereas p.Asp260fs failed to show any measurable hydrolytic activity toward oseltamivir. Acknowledging the limitations of in vitro methodologies, this fundamental alteration in the catalytic activity of CES1 strongly suggests that the activation of oseltamivir would be compromised in patients who express such CES1 variants. In addition to these two mutations, several other natural nonconservative CES1 variants were recently determined to also have functional significance (Shi et al., 2006; Tang et al., 2006). Furthermore, beyond the coding area mutations, a number of functional variants have been reported in the transcriptional regulation region of CES1 gene (Geshi et al., 2005; Hosokawa et al., 2008; Yoshimura et al., 2008). Among those, a single nucleotide polymorphism, –816A/C of the CES1A2 gene was found to be associated with an improved therapeutic response to an angiotensin-converting enzyme inhibitor imidapril, which is a prodrug and selectively activated by CES1 (Geshi et al., 2005).
Enzymatic kinetics study of oseltamivir hydrolysis catalyzed by WT CES1 and its variant p.Gly143Glu. The hydrolysis of oseltamivir (10–5000 μM) was determined after incubation with the cell S9 fractions at 37°C for 10 min. The Vmax and Km values were calculated using nonlinear regression analysis with GraphPad Prism software. Data present means ± S.D. for four independent experiments.
It was noted that the observed Vmax value of WT CES1 is consistent with that reported by Shi et al. (2006), whereas the Km value is seven times higher. We suspect this difference is more likely than not the result of different experimental conditions used in these two independent studies. For example, the reaction buffer used in the present study was phosphate-buffered saline containing 10 mM HEPES (pH 7.4), whereas a Tris buffer was used in the study by Shi et al. (2006). Indeed, a recent in vitro study addressing this very issue indicates that different enzymatic activity of CES could be observed when different assay buffers were used (Williams et al., 2008). Finally, the S9 fractions used in the present study were prepared from a stable CES1 cell line rather than a transient expression assay.
Functional CES1 is not only critical for the conversion of oseltamivir to its active metabolite to achieve a favorable therapeutic response, but it is also related to the toxicity during oseltamivir therapy. Converging evidence suggests that CES1 function in juvenile animals remain at a significantly lower level than that of adult animals (Kadner et al., 1992; Morgan et al., 1994; Moser et al., 1998; Karanth and Pope, 2000; Padilla et al., 2004; Anand et al., 2006). Animal studies demonstrated that juvenile rats did not hydrolyze oseltamivir efficiently, and they are more susceptible than adults to oseltamivir toxicity (http://www.fda.gov/cder/foi/nda/2000/21-246_Tamiflu_Pharmr.pdf). The present study suggests that, in addition to age, genetic variation is potentially an important factor influencing the enzymatic function of CES1 and could play a role in the therapeutic outcome and toxicity of pharmacotherapy with oseltamivir as well as other known CES1 substrates. Our previously published data with the psychostimulant methylphenidate indicate that the effects of CES1 variants on drug disposition are already advanced beyond the realm of speculation.
In summary, two newly identified CES1 mutations p.Gly143Glu and p.Asp260fs were determined to be dysfunctional enzymes with respect to the activation of the prodrug oseltamivir. Impaired enzymatic function could have significant implications with regard to both the therapeutic efficacy and tolerability of oseltamivir. It should be noted that the extremely low prevalence of p.Asp260fs mutation relegates its clinical significance to being very minor even though it results in a nonfunctional enzyme. However, p.Gly143Glu is a common variant in all populations assessed thus far, with the exception of Asians. A clinical study, particularly one assessing patients who have been genotyped and found to be heterozygously expressing p.Gly143Glu, is warranted to elucidate the influence of the p.Gly143Glu mutation on the pharmacological disposition and potential toxicities of oseltamivir.
Footnotes
-
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
-
doi:10.1124/dmd.108.024943.
-
ABBREVIATIONS: CES1, carboxylesterase 1; CES2, carboxylesterase 2; WT, wild type; PNPA, p-nitrophenyl acetate; PNP, p-nitrophenol; HPLC, high-performance liquid chromatography.
- Received September 28, 2008.
- Accepted November 18, 2008.
- The American Society for Pharmacology and Experimental Therapeutics