Abstract
UDP-Glucuronosyltransferases (UGTs) are phase II biotransformation enzymes that glucuronidate numerous endobiotic and xenobiotic substrates. Glucuronidation increases the water solubility of the substrate and facilitates renal and biliary excretion of the resulting glucuronide conjugate. UGTs have been divided into two gene families, UGT1 and UGT2. Tissue distribution of UGTs has not been thoroughly examined, and such data could provide insight into the importance of individual UGT isoforms in specific tissues and to the pharmacokinetics and target organ toxicity of UGT substrates. Therefore, the aim of this study was to determine mRNA levels of rat UGT1 and UGT2 family members in liver, kidney, lung, stomach, duodenum, jejunum, ileum, large intestine, cerebellum, and cerebral cortex, as well as nasal epithelium for UGT2A1. Tissue levels of UGT mRNA were detected using branched DNA signal amplification analysis. Three UGT isoforms, UGT1A1, UGT1A6, and UGT2B12, were detected in many tissues, whereas distribution of other UGT isoforms was more tissue-specific. For example, UGT2A1 was detected predominantly in nasal epithelium. Additionally, UGT1A5, UGT2B1, UGT2B2, UGT2B3, and UGT2B6 were detected primarily in liver. Furthermore, detection of UGT1A2, UGT1A3, UGT1A7, and UGT2B8 was somewhat specific to gastrointestinal (GI) tract. However, not all of these UGTs were detected in all portions of the GI tract. UGT1A8 was unique in that it was barely detectable in any of the tissues examined. In conclusion, some UGT isoforms were expressed in multiple tissues, whereas other UGT isoforms were predominantly expressed in a certain tissue such as nasal epithelium, liver, or GI tract.
UDP-Glucuronosyltransferases (UGTs2) are phase II biotransformation enzymes localized to the endoplasmic reticulum (Chowdhury et al., 1985). UGTs catalyze the conjugation of glucuronic acid to a broad spectrum of endobiotic and xenobiotic substrates with diverse chemical structures. Endogenous substrates for UGTs include compounds such as bilirubin, bile acids, serotonin, thyroid hormone, biogenic amines, and steroid hormones. Additionally, UGTs have a large number of xenobiotic substrates, including fat-soluble vitamins, carcinogens, plant metabolites, environmental pollutants, as well as drugs such as acetaminophen, chloramphenicol, diethylstilbestrol, morphine, and salicylic acid (Dutton, 1980; Clarke and Burchell, 1987; Mackenzie and Rodbourn, 1990;Burchell et al., 1991).
Based on sequence similarity, UGTs have been divided into two families, UGT1 and UGT2. All UGT1 family members are encoded from a single gene that consists of multiple first exons and four common exons (exons II, III, IV, and V). Individual UGT1A isoforms are formed by the splicing of one of the first exons to the four common exons, II to V. Distinct promoter regions have been identified for the multiple first exons, suggesting tissue-specific expression and inducer-responsive expression of individual UGT1A isoforms (Emi et al., 1995).
In rat, nine different first exons have been identified (Emi et al., 1995). Thus, the rat UGT1 family consists of nine members: UGT1A1, UGT1A2, UGT1A3, UGT1A4, UGT1A5, UGT1A6, UGT1A7, UGT1A8, and UGT1A9. However, rat UGT1A4 and UGT1A9 do not code for functional protein and are termed pseudogenes (Emi et al., 1995).
In contrast to the UGT1 family, UGT2 family members are encoded from individual genes, with each gene containing six exons (Mackenzie and Rodbourn, 1990; Haque et al., 1991). The UGT2 family has been further divided into two subfamilies, UGT2A and UGT2B. The rat 2A subfamily consists of a single olfactory-specific UGT, UGT2A1. The rat 2B subfamily consists of six members: UGT2B1, UGT2B2, UGT2B3, UGT2B6, UGT2B8, and UGT2B12. Members of the UGT2B subfamily have been named in the order they were cloned, regardless of species (Parkinson, 2001).
Glucuronidation increases the water solubility of the UGT substrate, which enhances urinary and biliary excretion of the resulting glucuronide conjugate. Glucuronidation is generally considered a detoxification reaction that terminates the biological activity of the substrate and facilitates its elimination from the body (Mulder, 1992). However, glucuronidation can result in the production of metabolites that are biologically active and/or toxic. For example, two metabolites of morphine, morphine-6-glucuronide and morphine-3-glucuronide, are biologically active. Morphine-6-glucuronide is a more potent analgesic than morphine itself (Paul et al., 1989), whereas morphine-3-glucuronide is an extremely effective antagonist of morphine analgesia (Smith et al., 1990).
Additionally, some glucuronide metabolites are toxic. For instance, the steroid hormone conjugates estradiol-17-β-d-glucuronide and testosterone-17-β-d-glucuronide have been shown to produce cholestasis (Meyers et al., 1980, 1981). Furthermore, acyl glucuronides of some nonsteroidal anti-inflammatory drugs, including zomepirec, ketoprofen, and diclofenac, can covalently bind to proteins after hydrolysis or rearrangement (Bailey and Dickinson, 1996; King et al., 2001).
Glucuronidation has been thought to occur primarily in liver; however, glucuronidation in extrahepatic tissues may have a significant impact on the pharmacokinetics and bioavailability of UGT substrates. For example, UGT isoforms present in intestine may contribute to the first-pass effect of some drugs. The capacity of a tissue to glucuronidate a substrate depends on the UGT isoforms present in the tissue and their abundance (Mackenzie and Rodbourn, 1990).
Tissue distribution of the rat UGT1 and UGT2 isoforms has not been fully characterized and could play an important role in determining target organ toxicity of UGT substrates. Tissue distribution of the rat UGT1A family has been examined in liver, kidney, and gastrointestinal tract (Emi et al., 1995; Grams et al., 2000). However, tissue distribution of the rat UGT2 family has been examined less thoroughly than the UGT1A family. Additionally, gender differences in rat UGT isoforms have not been thoroughly examined. Therefore, the purpose of this study was to quantitatively determine tissue- and gender-specific mRNA expression of rat UGT1 and UGT2 isoforms in liver, kidney, lung, stomach, duodenum, jejunum, ileum, large intestine, cerebellum, and cerebral cortex, as well as nasal epithelium for UGT2A1.
Materials and Methods
Animals.
Male and female Sasco Sprague-Dawley rats (200–250 g) were purchased from Charles River Laboratories, Inc. (Wilmington, MA). Animals were housed according to the American Animal Association Laboratory Animal Care guidelines. The rats were fed Laboratories Rodent Chow W (Harlan Laboratories, Madison, WI) and water ad libitum. Animals were decapitated and tissues, with the exception of intestine, were removed and snap-frozen in liquid nitrogen. The small intestine was sectioned into thirds; each section was dissected longitudinally and rinsed in saline. Intestinal epithelium was scraped from each section and snap-frozen in liquid nitrogen. Large intestine was similarly dissected, rinsed, and scraped. Tissues were stored at −80°C.
Tissues.
Tissues examined in the present tissue distribution study were chosen based on a preliminary screen in which pooled RNA samples (five animals per gender per tissue) were used to obtain single determinations of mRNA levels of UGT isoforms in the following 36 tissues: liver, kidney, lung, stomach, duodenum, jejunum, ileum, large intestine, cerebellum, cerebral cortex, heart, blood vessel, spleen, pancreas, thymus, muscle, skin, adrenal, lymph node, thyroid, eye, pituitary, thalamus, brain stem, caudate, frontal cortex, hippocampus, olfactory bulb, nasal epithelium, spinal cord, urinary bladder, testes, ventral prostate, dorsal prostate, ovary, and uterus (data not shown). From these data, liver, kidney, lung, stomach, duodenum, jejunum, ileum, large intestine, cerebellum, and cerebral cortex were selected as the major tissues of expression. Additionally, nasal epithelium was selected as a major tissue of expression in analysis of UGT2A1.
RNA Extraction.
Total tissue RNA was extracted using RNA-zolB reagent (Tel-Test Inc., Friendswood, TX) according to the manufacturer's protocol and resuspended in water treated with diethyl pyrocarbonate. RNA samples were analyzed by formaldehyde-agarose gel electrophoresis and integrity was confirmed by visualization of intact 18S and 28S rRNA.
Branched DNA Signal Amplification (bDNA) Assay.
UGT mRNA was measured using the bDNA assay (QuantiGene bDNA signal amplification kit; Bayer Diagnostics, East Walpole, MA) with modifications (Hartley and Klaassen, 2000). Rat UGT gene sequences of interest were acquired from GenBank. Multiple oligonucleotide probe sets [capture extender (CE), label extender (LE), and blocker (BL) probes] were designed using Probe Designer software, version 1.0, to be specific to a single mRNA transcript. The probes were designed with a Tm of approximately 63°C, enabling hybridization conditions to be held constant (i.e., 53°C) during each hybridization step and for each probe set. All probes designed in Probe Designer were submitted to the National Center for Biotechnological Information for nucleotide comparison by the basic logarithmic alignment search tool (BLASTn), to ensure minimal cross-reactivity with other known rat sequences and expressed sequence tags. Oligonucleotides with a high degree of similarity (≥80%) to other rat gene transcripts were eliminated from the probe set. Probe sets for UGT1A1, 1A2, 1A5, 1A6, 1A7, 2A1, and 2B1 were described previously (Vansell and Klaassen, 2002). New probe sets were designed for UGT1A3, 1A8, 2B2, 2B3, 2B6, 2B8, and 2B12 in an effort to improve signal by including BL probes and increasing the number of CE and LE probes. The new probes designed for UGT2B2 were added to the previous probe set. The nucleotide sequence of each new probe set is listed in Table1. Probe sets were not designed for UGT1A4 or UGT1A9, which are pseudogenes and were not examined in the present study.
Oligonucleotide probes for bDNA analysis of UGT mRNA levels
Total RNA (1 μg/μl; 5 μl/well; n = 4 or 5/tissue/gender) was added to the wells of a 96-well plate that contained 50 μl of capture hybridization buffer and 50 μl of diluted probe set (CE, LE, and BL probes). Total RNA was allowed to hybridize to the probe set overnight at 53°C. The mRNA transcript of interest is captured by hybridization with CE probes that have also hybridized with oligonucleotides fixed to the bottom of the wells of a 96-well plate. Subsequent hybridization steps result in hybridization of LE probes to the mRNA transcript of interest and to bDNA molecules. The branches of the bDNA molecules hybridized with alkaline phosphatase-conjugated oligonucleotides. Addition of an alkaline phosphatase substrate results in generation of chemiluminescence. Luminescence was measured with a Quantiplex 320 bDNA luminometer interfaced with Quantiplex Data Management software, version 5.02, for analysis of luminescence from 96-well plates. The luminescence for each well is reported as relative light units (RLUs) per 5 μg of total RNA.
Statistics.
Statistical differences were determined using Student's ttest with significance set at p ≤ 0.05.
Results
Preliminary Screen.
The present tissue distribution study was based on a preliminary screen in which pooled RNA samples (five animals per gender per tissue) were used to obtain single determinations of mRNA levels of UGT isoforms in 36 tissues of male and female Sprague-Dawley rats (data not shown; seeMaterials and Methods for a list of tissues). The results from the preliminary screen were used to determine the tissues that were examined in the present study. Tissues selected as major tissues of expression include liver, kidney, lung, stomach, duodenum, jejunum, ileum, large intestine, cerebellum, and cerebral cortex, as well as nasal epithelium for UGT2A1.
Tissue Distribution of the UGT1A Family.
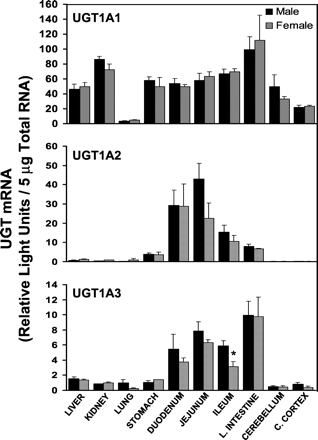
The tissue distribution of UGT1A1, UGT1A2, and UGT1A3 mRNA in male and female rats is shown in Fig. 1. UGT1A1 mRNA expression was detected in nearly all tissues examined. Tissue mRNA levels were similar in all tissues with the exception of lung in which mRNA levels were relatively low. Gender differences in UGT1A1 mRNA levels were not observed in any tissue examined.
Messenger RNA levels of UGT1A1, UGT1A2, and UGT1A3 in various tissues of rats (n = 4–5/gender) were determined by bDNA analysis and are expressed as RLUs/5 μg of total RNA.
Values are the mean ± S.E.M. ∗, significantly different from males (p ≤ 0.05).
In contrast to the somewhat ubiquitous distribution of UGT1A1, UGT1A2 mRNA was detected primarily in gastrointestinal tract. UGT1A2 mRNA levels were minimal in stomach, highest in duodenum and jejunum, and declined from jejunum to large intestine. No significant gender differences were observed in tissue UGT1A2 mRNA levels.
Similar to UGT1A2, UGT1A3 mRNA was primarily detected in the gastrointestinal tract, specifically in small and large intestine. However, the pattern of distribution differed from that of UGT1A2. In contrast to UGT1A2, UGT1A3 mRNA levels were more evenly distributed throughout the small intestine and highest in large intestine. A statistical difference was observed between male and female UGT1A3 mRNA levels in ileum, but the biological significance of this difference is uncertain. Although there is a tendency for females to have lower levels of UGT1A3 mRNA in intestine, the observed statistical difference in ileum is suspected to be artifactual given the lack of a clear sex difference in UGT1A3 mRNA levels in other sections of the intestine.
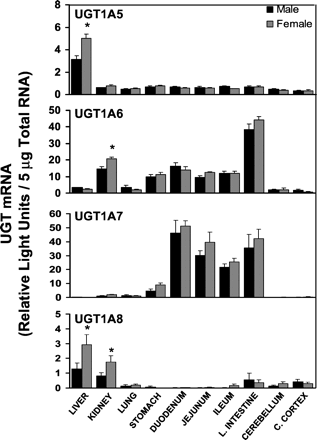
The distribution of UGT1A5, UGT1A6, UGT1A7, and UGT1A8 mRNA in male and female rat tissues is illustrated in Fig.2. Unlike other UGT1 family members, UGT1A5 mRNA expression seems to be limited to liver. Liver was the only tissue in which appreciable mRNA levels of UGT1A5 were detected. Additionally, UGT1A5 mRNA levels were approximately 35% higher in liver of female rats than that of male rats.
Messenger RNA levels of UGT1A5, UGT1A6, UGT1A7, and UGT1A8 in various tissues of rats (n = 4–5/gender) were determined by bDNA analysis and are expressed as RLUs/5 μg of total RNA.
Values are the mean ± S.E.M. ∗, significantly different from males (p ≤ 0.05).
Tissue distribution of UGT1A6 was similar to that of UGT1A1 in that both isoforms were detected in numerous tissues. UGT1A6 mRNA levels were highest in the large intestine and kidney, followed by stomach and small intestine. In contrast to UGT1A1 mRNA, which only displayed low levels in lung, UGT1A6 mRNA was detected at low levels in the liver, lung, cerebellum, and cerebral cortex compared with other tissues examined. Additionally, UGT1A6 mRNA levels were approximately 30% higher in kidney of female rats compared with male rats.
Tissue distribution of UGT1A7 was similar to UGT1A2 and UGT1A3, in that UGT1A7 mRNA was primarily detected in gastrointestinal tract. Low levels of UGT1A7 mRNA were detected in stomach. Highest levels were detected in duodenum and decreased to some extent through the small intestine. UGT1A7 mRNA levels in large intestine were similar to that of small intestine. UGT1A7 mRNA was also detected in lung and kidney, but at relatively low levels.
The last UGT1A family member, UGT1A8, was barely detectable in any tissue. Of the tissues examined, only liver and kidney exhibited mRNA levels greater than 1 RLU. Although UGT1A8 expression was very low compared with other UGT isoforms, gender differences were observed in both liver and kidney. In both tissues, mRNA levels in females were higher than in males.
Tissue Distribution of the UGT2 Family.
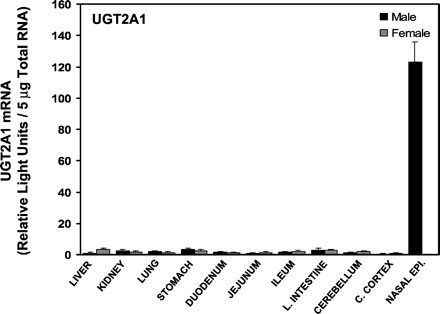
The tissue distribution of UGT2A1 mRNA is shown in Fig.3. UGT2A1 has previously been described as an olfactory-specific UGT (Lazard et al., 1991), and a preliminary study indicated nasal epithelium was a major tissue of expression. Therefore, for UGT2A1, nasal epithelium was included in the tissues examined. Moreover, UGT2A1 mRNA was predominantly detected in nasal epithelium. Potential gender differences were examined in all tissues with the exception of nasal epithelium, in which UGT2A1 mRNA levels were only examined in male rats. However, no gender differences were observed.
Messenger RNA levels of UGT2A1 in various tissues of rats (n = 4–5/gender) were determined by bDNA analysis and are expressed as RLUs/5 μg of total RNA.
Values are the mean ± S.E.M. ∗, significantly different from males (p ≤ 0.05).
Figure 4 illustrates the distribution of UGT2B1, UGT2B2, and UGT2B3 mRNA in male and female rat tissues. UGT2B1 and UGT2B2 mRNA were detected predominantly in liver. A similar gender difference was observed for both UGT2B1 and UGT2B2, in that liver mRNA levels of female rats were more than twice that of male rats. Similar to UGT2B1 and UGT2B2, UGT2B3 mRNA was mainly detected in liver, although no gender difference was observed. In contrast to distribution of UGT2B1 and UGT2B2 mRNA, low levels of UGT2B3 mRNA were also detected in duodenum and jejunum.
Messenger RNA levels of UGT2B1, UGT2B2, and UGT2B3 in various tissues of rats (n = 4–5/gender) were determined by bDNA analysis and are expressed as RLUs/5 μg of total RNA.
Values are the mean ± S.E.M. ∗, significantly different from males (p ≤ 0.05).
The tissue distribution of UGT2B6, UGT2B8, and UGT2B12 mRNA in male and female rats is illustrated in Fig. 5. Distribution of UGT2B6 mRNA was similar to UGT2B3. As with UGT2B3, UGT2B6 mRNA was detected mainly in liver, but at a low level. Like UGT2B3, no gender differences were observed in UGT2B6 mRNA levels. In contrast to other rat UGT isoforms in the 2B subfamily, which are primarily expressed in liver, UGT2B8 mRNA was mainly detected in stomach, duodenum, and kidney. In both stomach and duodenum, UGT2B8 mRNA levels were significantly higher in females than in males. Unlike other UGT2B isoforms, UGT2B12 was detected in all tissues examined. Highest levels of UGT2B12 mRNA were detected in liver and kidney. In intestine, UGT2B12 mRNA levels decreased from duodenum to large intestine. Although UGT2B12 mRNA was detected in all tissues examined, mRNA levels in ileum, large intestine, cerebral cortex, and cerebellum were very low in relation to liver and kidney levels. Gender differences in UGT2B12 mRNA levels were not observed in any tissue.
Messenger RNA levels of UGT2B6, UGT2B8, and UGT2B12 in various tissues of rats (n = 4–5/gender) were determined by bDNA analysis and are expressed as RLUs/5 μg of total RNA.
Values are the mean ± S.E.M. ∗, significantly different from males (p ≤ 0.05).
Discussion
Glucuronidation is a major pathway of metabolism for numerous endogenous and xenobiotic compounds. Thus, UGTs may have a significant impact on pharmacokinetics, bioavailability, and target organ toxicity of numerous compounds. The present study examined mRNA levels of UGT isoforms in various tissues of male and female rats in effort to fully characterize tissue-specific and gender-specific mRNA expression of UGT1 and UGT2 isoforms in rat. Tissue mRNA levels of UGTs were examined using the bDNA assay.
The bDNA signal amplification method has advantages over more commonly used methods to quantify mRNA in that it uses multiple short oligonucleotides as components of a larger probe set that retains the specificity to discriminate between closely related members of the same gene family without sacrificing sensitivity (Hartley and Klaassen, 2000). For instance, Northern blots use large sections of a cDNA as probes, whereas the bDNA assay uses short oligonucleotides as probes. Additionally, Northern blots rely upon quantification of an autoradiograph, whereas the bDNA assay directly measures luminescence, which is expressed as a numeric value that correlates to the amount of mRNA present. In contrast to reverse transcription-polymerase chain reaction, the bDNA assay works through noncycling, linear amplification, thus eliminating the chance for exponential amplification of a nonspecific binding event.
Several UGTs isoforms were expressed in liver. This was reasonable given that a large amount of glucuronidation occurs in liver. Surprisingly, of the seven members of the UGT1A family, only UGT1A1, UGT1A5, and UGT1A6 were detected at an appreciable level in liver. These findings confirm the results of a previous study that also detected these three transcripts in liver (Grams et al., 2000). However, the aforementioned study by Grams et al. (2000) also detected very low levels of UGT1A7 transcripts in liver. Yet, other studies have determined that UGT1A7 mRNA was undetectable in naive rat liver (Emi et al., 1995; Grove et al., 1997). In the present study, UGT1A8 mRNA was also detected in liver, but at levels barely above the detection limit. Previous studies have not detected UGT1A8 in any tissue examined, including liver (Emi et al., 1995; Grams et al., 2000). It is possible that both UGT1A7 and UGT1A8 are present in rat liver but at very low copy number, making their transcripts particularly difficult to detect.
In contrast to the rat UGT1 family, several rat UGT2B isoforms were detected in liver, including UGT2B1, UGT2B2, UGT2B3, UGT2B6, and UGT2B12. These isoforms have previously been identified in liver (Mackenzie, 1987, 1990; Haque et al., 1991; Green et al., 1995). Liver was the predominant tissue of expression for many UGT2B isoforms, including UGT2B1, UGT2B2, UGT2B3, and UGT2B6. UGT2B12 had high expression in liver but unlike other 2B isoforms was also expressed in multiple tissues. In contrast to other UGT2B isoforms, UGT2B8 mRNA levels in liver were negligible. UGT isoforms present in liver may contribute to the first-pass effect of many therapeutic drugs. Additionally, some of these UGTs may contribute to the generation of liver injury and/or cholestasis by glucuronidating steroid hormones such as endogenous estradiol, and exogenous xenobiotics, including diclofenac and related nonsteroidal anti-inflammatory drugs (Meyers et al., 1980; King et al., 2001).
In contrast to liver where many UGT2B isoforms were predominantly expressed, in the intestine, many UGT1 isoforms were expressed. Multiple UGT1 family members were expressed in intestine, including UGT1A1, UGT1A2, UGT1A3, UGT1A6, and UGT1A7. Of these UGT isoforms, UGT1A2, UGT1A3, and UGT1A7 were predominantly expressed in intestine. A previous study reported detection of UGT1A1, UGT1A2, UGT1A3, UGT1A6, and UGT1A7 mRNA in small intestine (Grams et al., 2000), in addition to UGT1A1, UGT1A2, UGT1A6, and UGT1A7, but not UGT1A3 mRNA in stomach and large intestine (Grams et al., 2000). However, in the present study, UGT1A3 mRNA was detected in both small and large intestine. This discrepancy could be due to differences in tissue collection or the methodology used to detect mRNA.
In contrast to the UGT1 family, few UGT2B subfamily members were detected in intestine. The existence of UGT2B1 and UGT2B3 in small intestine has been examined previously (Mackenzie, 1987). Mackenzie (1987) determined that both UGT2B1 and UGT2B3 mRNA were present at very low levels in small intestinal mucosa. The present study confirms the presence of UGT2B3 mRNA in small intestine but differs from the previous study in that UGT2B1 mRNA was not detected in small intestine. In the present study, two additional UGT2B members, UGT2B8 and UGT2B12, were detected in small intestine. UGT isoforms expressed in gastrointestinal tract may play a role in the first-pass effect of many therapeutic drugs administered orally and other chemicals, including morphine, 1-napthol, nalorphine, and harmol (Koster et al., 1985; Goon and Klaassen, 1991; Iwamoto and Klaassen, 1977a,b)
A few UGTs were expressed in multiple tissues. Of the 14 UGTs examined, only UGT1A1, UGT1A6, and UGT2B12 displayed this pattern of expression. These UGTs may play a significant role in detoxification of xenobiotics that have a tropism for extrahepatic tissues such as kidney, lung, or brain. In kidney, in addition to UGT1A1, UGT1A6, and UGT2B12, mRNA of UGT1A7 and UGT2B8 was detected but at low levels. The presence of UGT1A1, UGT1A6, UGT2B12, and low levels of UGT1A7 mRNA in kidney is in agreement with previous studies (Emi et al., 1995; Green et al., 1995;Grams et al., 2000; Auyeung et al., 2001). Mackenzie (1987) detected UGT2B1 and UGT2B3 mRNA in kidney at very low levels. However, this was not seen in the present study.
In both lung and brain, mRNA levels of UGT1A1, UGT1A6, and UGT2B12 were essentially very low, although both of these tissues have been shown to have UGT activity. A possible reason for such low mRNA levels in these tissues may be that the UGTs are expressed in a specific cell type that is either few in number or possibly limited to a specific region of the tissue. In a prior study, UGT1A6 was detected in rat brain and primary cultures of neurons and astrocytes (Suleman et al., 1998). UGT1A6 has also previously been detected in lung (Munzel et al., 1994). In the present study, low levels of UGT1A7 mRNA were detected in lung in addition to UGT1A1, UGT1A6, and UGT2B12. Tissue mRNA levels of UGT1A1, UGT1A6, and UGT1A7 have previously been examined in lung, however, only low levels of UGT1A6 and UGT1A7 mRNA were detected (Emi et al., 1995).
In the present study, tissue distribution of both male and female rats was examined in 10 tissues to determine potential gender differences in mRNA levels of the both UGT1 and UGT2 isoforms. Emi et al. (1995)examined sex differences in liver mRNA levels of rat UGT1A family members and found no remarkable sex difference. In general, the present study did not detect marked gender differences in either rat UGT1 or UGT2 families. However, some UGT isoforms were detected at higher levels in female rats. This was observed in more than one tissue, but mainly in liver. In the UGT1A family, UGT1A5 mRNA levels in liver of female rats were approximately 35% higher than in male rats. UGT1A6 mRNA levels in kidney of female rats were approximately 30% higher than in male rats. Even though UGT1A8 was barely detectable, mRNA levels in liver and kidney of female rats were significantly higher than in male rats.
As for the UGT2 family, UGT2B1 mRNA levels in liver of female rats were approximately 50% higher than that in male rats. UGT2B2 mRNA levels in liver of female rats were approximately 60% higher than in males. Gender differences in UGT2B2 expression were examined in a previous study but were not discernible (Haque et al., 1991). Gender differences in expression of UGT2B8 were seen in stomach and duodenum. UGT2B8 mRNA levels in stomach and duodenum of female rats were approximately 70 to 75% higher than in males.
In summary, the present study examined tissue-specific and gender-specific mRNA expression of rat UGT1 and UGT2 family members. UGT isoforms in liver include UGT1A1, UGT1A5, UGT1A6, UGT2B1, UGT2B2, UGT2B3, UGT2B6, UGT2B12, and possibly UGT1A8. UGT isoforms in intestine include UGT1A1, UGT1A2, UGT1A3, UGT1A6, UGT1A7, UGT2B3, UGT2B8, and UGT2B12. UGT isoforms in kidney include UGT1A1, UGT1A6, UGT1A7, UGT2B8, and UGT2B12. UGT isoforms in lung include UGT1A1, UGT1A6, UGT1A7, and UGT2B12. UGT isoforms in brain include UGT1A1, UGT1A6, and UGT2B12. There were not marked gender differences in the expression of UGT isoforms, however it was interesting that mRNA of some UGT isoforms were detected at higher levels in various tissues of female rats. These differences may play a role in gender-specific toxicity, gender differences in bioavailability, and pharmacokinetics of UGT substrates.
Acknowledgments
We thank Susan Buist, Dr. Tyra Leazer, Ning Li, and Dr. Angela Slitt for technical assistance.
Footnotes
-
↵1 Current address: Wyeth-Ayerst Research, 641 Ridge Rd., Chazy, NY 12121.
-
This study was supported by the National Institutes of Health Grant ES-03192. M.K.S. and N.R.V. were supported by National Institutes of Health Grant ES-07079.
- Abbreviations used are::
- GI
- gastrointestinal
- UGT
- uridine diphosphate glucuronosyltransferase
- bDNA
- branched deoxyribonucleic acid
- CE
- capture extender
- LE
- label extender
- BL
- blocker
- RLU
- relative light unit
- Received August 22, 2002.
- Accepted November 25, 2002.
- The American Society for Pharmacology and Experimental Therapeutics