Abstract
Human arylacetamide deacetylase (AADAC) is a major esterase responsible for the hydrolysis of clinical drugs such as flutamide, phenacetin, and rifampicin. Thus, AADAC is considered to be a relevant enzyme in preclinical drug development, but there is little information about species differences with AADAC. This study investigated the species differences in the tissue distribution and enzyme activities of AADAC. In human, AADAC mRNA was highly expressed in liver and the gastrointestinal tract, followed by bladder. In rat and mouse, AADAC mRNA was expressed in liver at the highest level, followed by the gastrointestinal tract and kidney. The expression levels in rat tissues were approximately 7- and 10-fold lower than those in human and mouse tissues, respectively. To compare the catalytic efficiency of AADAC among three species, each recombinant AADAC was constructed, and enzyme activities were evaluated by normalizing with the expression levels of AADAC. Flutamide and phenacetin hydrolase activities were detected by the recombinant AADAC of all species. In flutamide hydrolysis, liver microsomes of all species showed similar catalytic efficiencies, despite the lower AADAC mRNA expression in rat liver. In phenacetin hydrolysis, rat liver microsomes showed approximately 4- to 6.5-fold lower activity than human and mouse liver microsomes. High rifampicin hydrolase activity was detected only by recombinant human AADAC and human liver and jejunum microsomes. Taken together, the results of this study clarified the species differences in the tissue distribution and enzyme activities of AADAC and facilitate our understanding of species differences in drug hydrolysis.
Introduction
Enzymatic drug hydrolysis plays important roles in the metabolic activation and detoxification of clinically used drugs and prodrugs including ester, amide, and thioester bond. Although several esterases are known to catalyze drug metabolism, carboxylesterase (CES) is a representative serine esterase contributing to the hydrolysis of various drugs and xenobiotics. In human, CES1 and CES2 families have only two (CES1 and pseudogene CES1P1) and single genes, respectively, although the CES1P1 gene is altered to the functional CES1 gene in some persons (Fukami et al., 2008). Meanwhile, rat and mouse have multiple Ces1 and Ces2 genes (five and seven genes, respectively, in rat and eight genes for both in mouse) (Holmes et al., 2010). In addition, the tissue distribution and substrate specificity of CES are different between human and rodents (Luan et al., 1997; Sanghani et al., 2002; Satoh et al., 2002). Thus, studies on species differences in CES have been advanced.
In recent studies, we found that human arylacetamide deacetylase (AADAC), as well as CES enzymes, is involved in drug metabolism (Watanabe et al., 2009, 2010; Nakajima et al., 2011). Human AADAC is a major serine esterase expressed in liver and intestine (Probst et al., 1994). Rodents also have a single AADAC gene, and the amino acid identities between human and rodent (rat and mouse) AADAC are not high (68.2 and 69.9%, respectively). It was reported, using a limited number of tissues, that rat and mouse AADAC mRNA is expressed in liver and small intestine (Trickett et al., 2001). The function and tissue distribution of rodent AADAC are poorly understood.
Human AADAC is responsible for the hydrolysis of clinically relevant drugs such as flutamide, phenacetin, and rifamycins. Flutamide is a nonsteroidal antiandrogen drug for prostate cancer. Flutamide is mainly metabolized to 2-hydroxyflutamide, a pharmacologically active metabolite, by human CYP1A2, and is also hydrolyzed to 4-nitro-3-(trifluoromethyl)phenylamine (FLU-1) by AADAC (Katchen and Buxbaum, 1975; Schulz et al., 1988; Watanabe et al., 2009). N-Hydroxyl FLU-1, which is a metabolite of FLU-1 by human CYP3A4, has been suggested to be associated with hepatotoxicity (Goda et al., 2006). Although phenacetin had been widely used as an analgesic antipyretic, it was withdrawn from the market because of renal failure (Sicardi et al., 1991; Gago-Dominguez et al., 1999). Phenacetin is primarily metabolized to acetaminophen by CYP1A2 and is also hydrolyzed to p-phenetidine by human AADAC (Butler et al., 1989; Watanabe et al., 2010). p-Phenetidine is considered to be further metabolized to N-hydroxyphenetidine, the possible metabolite causing nephrotoxicity and hematotoxicity (Wirth et al., 1982; Jensen and Jollow, 1991). From these lines of evidence, it is considered that AADAC is associated with the occurrence of flutamide and phenacetin toxicities. Rifamycins such as rifampicin, rifabutin, and rifapentine have been used as antituberculosis drugs. Rifamycins have the potential to induce various drug-metabolizing enzymes, especially cytochrome P450s (Grange et al., 1994). In addition, rifampicin is suggested to be associated with hepatotoxicity by exacerbating isoniazid and/or other antituberculosis drug-induced hepatotoxicity or by direct toxic injury to hepatocytes (Gangadharam, 1986). However, we demonstrated that 25-deacetylrifamycins, the principal metabolites of rifamycins by AADAC in human, have lower induction potency and toxicity compared with rifamycins (Nakajima et al., 2011). Thus, AADAC seems to play a role in the attenuation of induction of drug-metabolizing enzymes and toxicity by rifamycins.
In drug development, experimental animals are frequently used to predict the excretory pathway and evaluate the toxicity of drug candidates, but sometimes species differences in drug-metabolizing enzymes make extrapolations to human difficult. Because AADAC is associated with drug toxicity as described above, investigation of species differences with AADAC will contribute to the interpretation of the data for humans and experimental animals. In the present study, we investigated the species differences in tissue distribution and enzyme activities of AADAC in human, rat, and mouse.
Materials and Methods
Chemicals and Reagents.
Flutamide, FLU-1, phenacetin, and rifampicin were purchased from Wako Pure Chemicals (Osaka, Japan). p-Phenetidine, aprotinin, bestatin, leupeptin, and trypsin inhibitor were purchased from Sigma-Aldrich (St. Louis, MO). 25-Desacetylrifampicin (25-deacetylrifampicin) was from Toronto Research Chemicals Inc. (Toronto, Canada). Primers were commercially synthesized at Hokkaido System Sciences (Sapporo, Japan). The random hexamer and SYBR Premix Ex Taq were from Takara (Shiga, Japan). RNAiso was from Nippon Gene (Tokyo, Japan). ReverTra Ace (Moloney murine leukemia virus reverse transcriptase RNaseH −) was obtained from Toyobo (Tokyo, Japan). All other chemicals used in this study were of analytical grade or the highest quality commercially available.
Animals.
Sprague-Dawley rats (6-week-old male, 160–180 g, and female, 120–140 g) and C57BL/6J mice (6-week-old male, 20–25 g, and female, 15–20 g) were obtained from SLC Japan (Hamamatsu, Japan). Animals were housed in the institutional animal facility in a controlled environment (temperature 25 ± 1°C and 12-h light/dark cycle) with access to food and water ad libitum. Animals were acclimatized for 1 week before use. Animals were maintained in accordance with the National Institutes of Health Guide for Animal Welfare of Japan, as approved by the Institutional Animal Care and Use Committee of Kanazawa University.
RNA Preparation from Rat and Mouse Tissues and Real-Time RT-PCR Analyses.
Total RNA samples from rat and mouse liver, kidney, stomach, jejunum (enterocytes), lung, trachea, thymus gland, adrenal gland (rat only), pancreas, heart, spleen, bladder, testis, epididymis adipose tissue (male rat only), prostate, ovary, and uterus were freshly isolated using RNAiso. Mouse adrenal gland was too small to be excised. Real-time RT-PCR was performed for quantitative determination of AADAC mRNA using an MX3000P real-time PCR system (Agilent Technologies, Santa Clara, CA). The forward and reverse primers used for PCR were rAADAC-RT-F and rAADAC-RT-R primers for rat or mAADAC-RT-F and mAADAC-RT-R primers for mouse (Table 1). A 1-μl portion of the reverse-transcribed mixture was added to a PCR mixture containing 10 pmol of each primer and 12.5 μl of SYBR Premix Ex Taq solution in a final volume of 25 μl. After an initial denaturation at 95°C for 3 min, the amplification was performed by denaturation at 95°C for 30 s, annealing at 54°C (rat) or 55°C (mouse) for 20 s, and extension at 72°C for 20 s for 45 cycles. The copy numbers were calculated using standard amplification curves, which were obtained using plasmids containing a full-length cDNA as a template.
Sequence of primers used in this study
Preparation of Tissue Microsomes.
Human liver microsomes (HLM) (pooled, n = 50) were purchased from BD Gentest (Woburn, MA). Human jejunum microsomes (HJM) (pooled, n = 10), human renal microsomes (HRM) (single donor), and human pulmonary microsomes (single donor) were purchased from Tissue Transformation Technologies (Edison, NJ).
Microsomes of pooled liver, jejunum, kidney, and lung were prepared from five male mice and three male rats according to the method of Emoto et al. (2000) with slight modifications. Liver, kidney, and lung were suspended in 3 volumes of ice-cold buffer A [50 mM Tris-HCl buffer (pH 7.4) containing 150 mM KCl, 20% (v/v) glycerol, 1 mM EDTA] and homogenized using a motor-driven Teflon-tipped pestle. The homogenate was centrifuged at 9000g for 15 min, and then the supernatant was centrifuged at 105,000g for 90 min. The microsomal pellets were resuspended in 3 volumes of ice-cold buffer A before resedimentation at 105,000g for 60 min. The microsomal fraction was resuspended in 1 volume of ice-cold buffer B [20% (v/v) glycerol and 250 mM sucrose] and homogenized. These procedures were performed at 4°C. Jejunums were divided, cut longitudinally, and then washed in ice-cold 1.15% KCl by gentle swirling, and mucosal cells were gently scraped off with a microspatula. The subsequent procedure was the same as for the other tissues except that buffer A included 1 mg/ml trypsin inhibitor, 10 μM leupeptin, 0.04 U/ml aprotinin, and 1 μM bestatin. The protein concentrations were determined according to the method of Bradford (1976) using γ-globulin as the standard.
Construction of Human, Rat, and Mouse Intact and Histidine-Tagged AADAC Expression Vectors.
To compare the enzyme activities between AADACs of three species, AADAC expression vectors with the intact cDNA sequence were constructed. The bacmid DNA containing human AADAC was constructed in our previous study (Watanabe et al., 2010). In this study, the expression plasmids for rat and mouse AADAC using a Bac-to-Bac Baculovirus Expression System (Invitrogen, Carlsbad, CA) were constructed according to the manufacturer's protocol. Rat and mouse AADAC cDNAs were prepared by the RT-PCR technique using total RNA from liver. The forward and reverse primers were rAADAC-F and rAADAC-R primers for rat or mAADAC-F and mAADAC-R for mouse (Table 1). The PCR products were subcloned into the pFastBac1 vector.
To normalize the reactivity of human AADAC antibody against human, rat, and mouse AADAC, His-tagged AADAC expression vectors were also constructed. AADAC cDNAs with the sequences encoding the His tag (5 histidines) just before the stop codon were prepared using the above pFastBac1 vector containing AADAC cDNA by PCR with the following primers: pFastBac1-F and hHis-AADAC-R for human, rHis-AADAC-F and rHis-AADAC-R for rat, or mHis-AADAC-F and mHis-AADAC-R for mouse (Table 1). The PCR products were ligated into the pFastBac1 vector using appropriate restriction enzymes.
The pFastBac1 vectors described above were transformed into DH10Bac competent cells, followed by transposition of the inserts into bacmid DNA. The sequence of the AADAC cDNA was determined using a Thermo Sequenase Cy5.5 Dye Terminator Cycle Sequencing kit (GE Healthcare, Chalfont St. Giles, Buckinghamshire, UK) with a Long-Read Tower DNA sequencer (GE Healthcare). In this study, the nucleotide sequences of rat and mouse AADAC are referred to as BC088143.1 and NM_023383.1, respectively. Nonrecombinant bacmid DNA (mock) was also prepared by the same procedures.
Expression of Human, Rat, and Mouse AADAC in Sf21 Cells.
Spodoptera frugiperda Sf21 cells (Invitrogen) were grown in Sf-900 II SFM containing 10% fetal bovine serum at 27°C. The recombinant and mock bacmid DNAs were separately transfected into Sf21 cells with Cellfectin Reagent (Invitrogen), and the virus was harvested by collecting the cell culture medium at 72 h after transfection. Cells were routinely harvested 7 days after infection, washed twice with phosphate-buffered saline, and stored at −80°C until analysis. Cell homogenates were prepared by suspension in TGE buffer [10 mM Tris-HCl buffer (pH 7.4), 20% glycerol, and 1 mM EDTA (pH 7.4)] and disruption by freeze-thawing three times according to our previous study (Watanabe et al., 2010). Then, the suspensions were homogenized with a Teflon-glass homogenizer for 10 strokes.
Immunoblot Analysis.
SDS-polyacrylamide gel electrophoresis and immunoblot analysis were performed according to Laemmli (1970). To examine the expression levels of His-tagged AADAC, Sf21 cell homogenates expressing His-tagged AADAC (20 μg) were separated on 10% polyacrylamide gels and electrotransferred onto a polyvinylidene difluoride membrane (Immobilon-P; Millipore Corporation, Billerica, MA). The membrane was probed with monoclonal mouse anti-tetra His antibody (QIAGEN, Valencia, CA) and the corresponding fluorescent dye-conjugated second antibody, and an Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE) was used for the detection. The band intensity was quantified using ImageQuant TL image analysis software (GE Healthcare). The relative expression level was determined by the band intensity.
Next, the relative reactivity of monoclonal mouse anti-human AADAC antibody (Abnova, Taipei City, Taiwan) against the recombinant AADAC of each species was determined. Homogenates of Sf21 cell-expressing His-tagged human, rat, and mouse AADAC (20, 41.7, and 7.6 μg, respectively), which show similar band intensities between three species by using anti-tetra His antibody, were loaded. The detection method was the same as that described above except that monoclonal mouse anti-human AADAC antibody was used. The relative reactivity of monoclonal mouse anti-human AADAC antibody against the AADAC of each species was determined by the band intensity.
After the reactivity of the AADAC antibody was evaluated, immunoblot analysis for recombinant intact AADAC was performed. Sf21 cell homogenates expressing recombinant, intact AADAC (20 μg) were loaded, and AADAC protein was detected with monoclonal mouse anti-human AADAC antibody. The band intensity was quantified and then normalized by the reactivity of the AADAC antibody. Thus, the relative expression level of recombinant intact AADAC was estimated. These immunoblot analyses were performed in the linear range of band intensity with respect to the amount of protein.
Flutamide, Phenacetin, and Rifampicin Hydrolase Activities.
Flutamide, phenacetin, and rifampicin hydrolase activities were determined using human, rat, and mouse tissue microsomes and Sf21 cell homogenates expressing recombinant AADAC. Flutamide hydrolase activity was determined according to Watanabe et al. (2009). A typical incubation mixture (final volume of 0.2 ml) contained 100 mM potassium phosphate buffer (pH 7.4) and various enzyme sources (microsomal proteins and Sf21 cell homogenates expressing AADAC: 0.4 mg/ml). In the preliminary study, we confirmed that the rate of FLU-1 formation was linear with respect to the protein concentrations (<0.6 mg/ml microsomal protein and Sf21 cell homogenates expressing AADAC) and incubation time (<45 min).
The phenacetin deacetylase (hydrolase) activity was determined according to Watanabe et al. (2010) with a slight modification. A typical incubation mixture (final volume of 0.2 ml) contained 100 mM potassium phosphate buffer (pH 7.4) and various enzyme sources (microsomal proteins and Sf21 cell homogenates expressing human AADAC: 0.4 mg/ml; rat and mouse AADAC: 0.2 mg/ml). In the preliminary study, we confirmed that the rate of formation of p-phenetidine was linear with respect to the protein concentrations (<1.0 mg/ml microsomal proteins and Sf21 cell homogenates expressing human and rat AADAC, and <0.2 mg/ml Sf21 cell homogenates expressing mouse AADAC) and incubation time (<60 min for microsomal protein and Sf21 cell homogenates expressing human AADAC, and <10 min for Sf21 cell homogenates expressing rat and mouse AADAC).
The rifampicin deacetylase (hydrolase) activity was determined according to Nakajima et al. (2011). A typical incubation mixture (final volume of 0.2 ml) contained 100 mM potassium phosphate buffer (pH 7.4) and various enzyme sources (microsomal protein and Sf21 cell homogenates expressing human, rat, and mouse AADAC: 0.5 mg/ml). In the preliminary study, we confirmed that the rate of formation of 25-deacetylrifampicin was linear with respect to the protein concentration (<1.5 mg/ml microsomal protein and Sf21 cell homogenates expressing AADAC) and incubation time (<90 min).
Each reaction was performed from three independent experiments. For the kinetic analyses of flutamide, phenacetin, and rifampicin hydrolase activity, the parameters were estimated from the fitted curves using a computer program (KaleidaGraph; Synergy Software, Reading, PA) designed for nonlinear regression analysis. The kinetic equations were obtained by fitting appropriate curves with the highest R2 values.
Results
Expression of AADAC mRNA in Human, Rat, and Mouse Tissues.
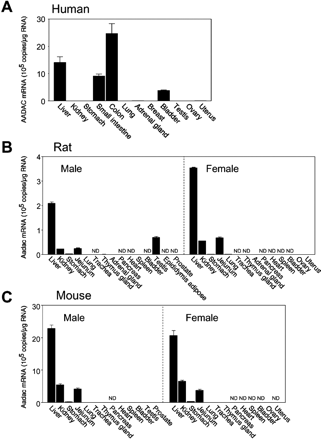
The expression levels of AADAC mRNA in human, rat, and mouse tissues were determined by real-time RT-PCR analysis (Fig. 1). The integrity of the extracted total RNA was verified by gel electrophoresis of 1 μg of RNA on a 0.8% agarose gel, which showed sharp and clear 28S and 18S rRNA bands (data not shown). The data for human AADAC mRNA were cited from our previous study (Watanabe et al., 2009). In human, AADAC mRNA was highly expressed in liver and the gastrointestinal tract and moderately expressed in bladder. In rat and mouse, AADAC mRNA was expressed in liver at the highest level, followed by the gastrointestinal tract (jejunum) and kidney. In most tissues, the expression levels in rat tissues were approximately 7- and 10-fold lower than those in human and mouse tissues, respectively. Approximately 1.5- to 3.0-fold higher expression of AADAC mRNA in female rat tissues was observed compared with that in male rat tissues. However, gender differences in AADAC expression level were not detected in mouse.
Expression levels of AADAC mRNA in various tissues of human (A), Sprague-Dawley rat (B), and C57BL/6J mouse (C). Copy numbers of AADAC mRNA were determined by real-time RT-PCR analyses. Each column represents the mean ± S.D. of triplicate determinations. ND, not detected.
Expression Levels of Recombinant Human, Rat, and Mouse AADACs in Sf21 Cells.
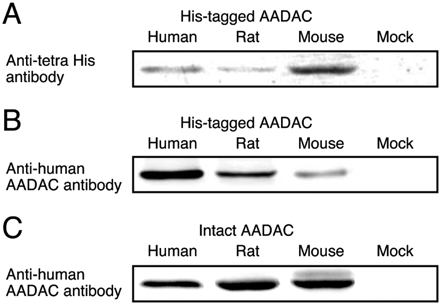
The expression levels of human, rat, and mouse AADAC in our expression systems were determined by immunoblot analysis using monoclonal mouse anti-human AADAC antibody. However, the reactivity of the antibody against AADAC for each species might be different. Therefore, Sf21 cells expressing His-tagged AADAC were established. Homogenates of Sf21 cells expressing His-tagged AADAC were loaded for SDS-PAGE and were reacted with monoclonal mouse anti-tetra His antibody (Fig. 2A). The relative band intensities (human, rat, and mouse = 1.00 ± 0.22, 0.48 ± 0.06, and 2.64 ± 0.17) reflect the relative expression levels of His-tagged AADAC.
A, representative photograph of immunoblot analysis of the expression level of His-tagged AADAC. Loaded Amounts of homogenates of Sf21 cells expressing His-tagged AADAC loaded were 20 μg. B, representative photograph of immunoblot analysis of the reactivity of antihuman AADAC antibody to AADAC from each species. Homogenates of Sf21 cells expressing His-tagged human, rat, and mouse AADAC (20, 41.7, and 7.6 μg, respectively), which showed similar band intensities with anti-tetra His antibody, were loaded. C, representative photograph of immunoblot analysis for determination of the expression level of recombinant intact AADAC protein. Loaded amounts of homogenates expressing recombinant intact AADAC of the three species were 20 μg. These immunoblot analyses were performed in triplicate.
According to the relative expression levels of His-tagged AADAC, homogenates of Sf21 cells expressing His-tagged human, rat, and mouse AADAC, which show similar band intensities with anti-tetra His antibody, were loaded. The membranes of His-tagged AADACs were reacted with anti-human AADAC antibody (Fig. 2B). The relative band intensities (human, rat, and mouse = 1.00 ± 0.13, 0.48 ± 0.11, and 0.16 ± 0.04) reflect the relative reactivity of anti-human AADAC antibody against the AADAC of each species.
Finally, intact AADAC proteins in homogenates of Sf21 cells were detected using anti-human AADAC antibody (Fig. 2C). The relative band intensities were as follows: human, rat, and mouse = 1.00 ± 0.14, 1.46 ± 0.21, and 1.31 ± 0.10. These values were divided by the relative reactivity of anti-human AADAC antibody against the AADAC of each species (Fig. 2B), resulting in the correct relative expression levels of recombinant intact AADAC in our expression systems. Given that the expression level of recombinant human AADAC was 1.00 unit/mg, the rat and mouse AADAC expression levels were 3.04 and 8.20 units/mg, respectively. In the subsequent study, the enzyme activities by the expression systems were normalized with these units.
Kinetic Analyses of Flutamide Hydrolase Activities by Recombinant AADACs and Tissue Microsomes of Human, Rat, and Mouse.
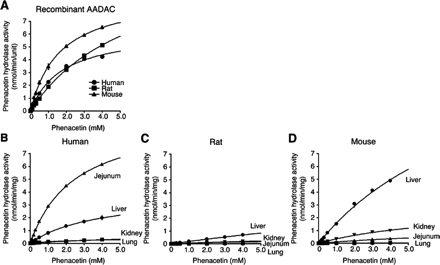
We found that AADAC is the principal enzyme in flutamide hydrolysis in human (Watanabe et al., 2009). The flutamide hydrolase activities by recombinant AADAC and tissue microsomes of human, rat, and mouse were analyzed (Fig. 3; Table 2). For some enzyme sources, the maximum substrate concentration (0.75 mM) was not sufficiently high to determine the Km values because of the limited solubility of flutamide in the incubation mixture. Therefore, the CLint values were calculated with the initial slope of the velocity versus the substrate concentration. In recombinant AADAC (Fig. 3A), the flutamide hydrolase activities were detected in rodents as well as in human and were fitted to the Michaelis-Menten equation. The Km, Vmax, and CLint values of human AADAC were 0.6 ± 0.1 mM, 1.1 ± 0.1 nmol · min−1 · unit−1, and 1.7 ± 0.0 μl · min−1 · unit−1, respectively. In recombinant rat AADAC, the Km and Vmax values were higher than those in human AADAC, resulting in a lower CLint value (1.0 ± 0.0 μl · min−1 · unit−1). Likewise, recombinant mouse AADAC showed higher Km and Vmax values, resulting in a CLint value (0.9 ± 0.0 μl · min−1 · unit−1) that was lower than that in human AADAC. Thus, human AADAC showed the highest catalytic efficiency for flutamide hydrolysis among the three species.
Kinetic analyses of flutamide hydrolase activities by AADAC expressed in Sf21 cells (A) and in four tissue microsomes of human (B), rat (C), and mouse (D). Total cell homogenates and microsomes were incubated with 0.025 to 0.75 mM flutamide (HJM were incubated with 0.005–0.75 mM flutamide). Each data point represents the mean ± S.D. of triplicate determinations.
Kinetic parameters for flutamide hydrolase activities by recombinant AADAC and tissue microsomes
Data represent the mean ± S.D. of triplicate determinations. CLint values of human AADAC and MLM are expressed as Vmax/Km.
Among human tissue microsomes (Fig. 3B), high flutamide hydrolase activities were detected in jejunum and liver microsomes, reflecting the high expression of AADAC mRNA in these tissues (Fig. 1A). In HRM, the activity was detected at low flutamide concentrations (Fig. 3B; Supplemental Fig. 1). Similar to those for recombinant human AADAC, the kinetics in HLM were fitted to the Michaelis-Menten equation. On the other hand, the kinetics in HJM and HRM were fitted to the combined Michaelis-Menten equation/substrate inhibition equation and the substrate inhibition equation, respectively. Thus, in HJM, two Km and Vmax values were calculated with the Michaelis-Menten equation (4.7 ± 2.1 mM and 7.8 ± 3.0 nmol · min−1 · mg−1, respectively) and substrate inhibition equation (0.2 ± 0.1 mM and 1.9 ± 0.6 nmol · min−1 · mg−1, respectively) (Table 2). In rat, although the activity by recombinant rat AADAC was fitted to the Michaelis-Menten equation, that in rat tissue microsomes was fitted to the Hill equation (Fig. 3C). High activity was detected in liver microsomes with a CLmax value of 1.4 ± 0.1 μl · min−1 · mg−1. In addition, the activities were also detected in rat jejunum microsomes, rat renal microsomes (RRM), and rat pulmonary microsomes with low CLmax values (0.1 ± 0.0–0.2 ± 0.0 μl · min−1 · mg−1). In mouse tissue microsomes (Fig. 3D), high activity was detected in liver microsomes with a CLint value of 1.2 ± 0.0 μl · min−1 · mg−1. The activities were also detected in mouse jejunum microsomes (MJM) and mouse renal microsomes (MRM), with CLint values of 0.2 ± 0.0 and 0.6 ± 0.0 μl · min−1 · mg−1, respectively. Similar to those in recombinant mouse AADAC, these activities in mouse liver microsomes (MLM), MJM, and MRM were fitted to the Michaelis-Menten equation. Mouse pulmonary microsomes (MPM) showed quite low activity. Thus, remarkable species differences in flutamide hydrolysis were observed in jejunum.
Kinetic Analyses of Phenacetin Hydrolase Activities by Recombinant AADACs and Tissue Microsomes of Human, Rat, and Mouse.
We found that AADAC is the principal enzyme responsible for phenacetin hydrolysis in human (Watanabe et al., 2010). The phenacetin hydrolase activities by recombinant AADAC and tissue microsomes of human, rat, and mouse were analyzed (Fig. 4; Table 3). For some enzyme sources, the maximum substrate concentration (4 mM) was not sufficiently high to determine the Km values because of the limited solubility of phenacetin in the incubation mixture. Therefore, the CLint values were calculated with the initial slope of the velocity versus the substrate concentration. In recombinant AADAC (Fig. 4A), the phenacetin hydrolase activities were detected in rodents as well as in human and were fitted to the Michaelis-Menten equation. The Km, Vmax, and CLint values of human AADAC were 1.8 ± 0.1 mM, 6.4 ± 0.2 nmol · min−1 · unit−1, and 3.5 ± 0.1 μl · min−1 · unit−1, respectively. In recombinant rat AADAC, the Km and Vmax values were higher than those in human AADAC, resulting in a lower CLint value (2.2 ± 0.0 μl · min−1 · unit−1). In contrast, recombinant mouse AADAC showed Km and Vmax values similar to those of human AADAC, resulting in a CLint value of 5.7 ± 0.3 μl · min−1 · unit−1. Thus, human and mouse AADAC showed higher catalytic efficiency than rat AADAC.
Kinetic analyses of phenacetin hydrolase activities by AADAC expressed in Sf21 cells (A) and in four tissue microsomes of human (B), rat (C), and mouse (D). Total cell homogenates and microsomes were incubated with 0.05 to 4 mM phenacetin. Each data point represents the mean ± S.D. of triplicate determinations.
Kinetic parameters of phenacetin hydrolase activities by recombinant AADAC and tissue microsomes
Data represent the mean ± S.D. of triplicate determinations. CLint values of human AADAC, HLM, HJM, HRM, mouse AADAC, and MPM are expressed as Vmax/Km.
In human tissue microsomes (Fig. 4B), high phenacetin hydrolase activities were detected in jejunum and liver microsomes, with CLint values of 4.0 ± 0.1 and 1.1 ± 0.0 μl · min−1 · mg−1, respectively. In rat tissue microsomes (Fig. 4C), liver microsomes showed relatively high activity, and the CLint value was similar to those in other tissue microsomes (0.1 ± 0.0–0.2 ± 0.0 μl · min−1 · mg−1) (Table 3) because of the high Km value in liver microsomes. In mouse tissue microsomes (Fig. 4D), high activity was detected in liver microsomes with a CLint value of 1.3 ± 0.1 μl · min−1 · mg−1, which was similar to that in HLM. The CLint values of other tissues (MJM, 0.1 ± 0.0 μl · min−1 · mg−1; MRM, 0.4 ± 0.1 μl · min−1 · mg−1; and MPM, 0.0 ± 0.0 μl · min−1 · mg−1) were lower than that of MLM. The phenacetin hydrolase activities in human, rat, and mouse tissues appeared to be correlated with the expression levels of AADAC mRNA (Fig. 1) and were fitted to the Michaelis-Menten equation, as was the case with those by recombinant AADAC.
Kinetic Analyses of Rifampicin Hydrolase Activities by Recombinant AADACs and Tissue Microsomes of Human, Rat, and Mouse.
We found that AADAC is the principal enzyme responsible for the rifampicin hydrolysis in human (Nakajima et al., 2011). The rifampicin hydrolase activities by recombinant AADAC and tissue microsomes of human, rat, and mouse were analyzed (Fig. 5; Table 4). In recombinant AADACs (Fig. 5A), human AADAC showed rifampicin hydrolase activity, which fitted the substrate inhibition equation. The Km, Vmax, and CLint values were 0.2 ± 0.0 mM, 149.0 ± 9.0 pmol · min−1 · unit−1, and 0.9 ± 0.1 μl · min−1 · unit−1, respectively. In contrast, rat and mouse AADAC did not show the activity.
Kinetic analyses of rifampicin hydrolase activities by AADAC expressed in Sf21 cells (A) and in four tissue microsomes of human (B), rat (C), and mouse (D). Total cell homogenates and microsomes were incubated with 0.005 to 1 mM rifampicin. Each data point represents the mean ± S.D. of triplicate determinations.
Kinetic parameters for rifampicin hydrolase activities by recombinant AADAC and tissue microsomes
Data represent the mean ± S.D. of triplicate determinations. CLint values of human AADAC, HLM, and HJM are expressed as Vmax/Km.
In human tissue microsomes (Fig. 5B), jejunum and liver microsomes showed high rifampicin hydrolase activity, which fitted the substrate inhibition equation. Although the activity in HJM seemed to be higher than that in HLM, the CLint values in both microsomes were similar (both tissue microsomes: 0.7 ± 0.0 μl · min−1 · mg−1). The Km values of liver and jejunum microsomes were 0.4 ± 0.0 and 0.3 ± 0.0 mM, respectively, which were similar to the Km value of human AADAC (0.2 ± 0.0 mM). In rat and mouse tissue microsomes (Fig. 5, C and D), the activities were too low to calculate the kinetic parameters (Table 4). These results suggested the species differences in the substrate specificity of AADAC.
Discussion
Our recent studies found that human AADAC is responsible for the hydrolysis of clinically relevant drugs such as flutamide, phenacetin, and rifamycins (Watanabe et al., 2009, 2010; Nakajima et al., 2011). Because these drugs were reported to cause adverse drug reactions, AADAC is a pharmacologically and toxicologically relevant enzyme. In drug development, a preclinical study is conducted for the prediction of clinical efficacy, metabolic pathway, and toxicity using laboratory animals. Therefore, an understanding of the species differences in drug metabolism is important to evaluate the efficacy and safety for humans. However, the species differences with AADAC have not been evaluated. The amino acid identities between human and rat or mouse AADAC are not high (68.2 and 69.9%, respectively) (Fig. 6), considering that there are species differences in the catalytic efficiency and substrate specificity of AADAC. In this study, we evaluated the tissue distribution and enzyme activities of human, rat, and mouse AADAC.
Alignment of the amino acid sequence of human, rat, and mouse AADAC. Identical amino acids between three species are shown in gray boxes. The presumed active site Gly-X-Ser-X-Gly motif and aspartic acid and histidine, which complete the catalytic triad, are shown in bold type.
At first, the tissue distribution and expression levels of AADAC in the three species were investigated (Fig. 1). In all species, AADAC mRNA was highly expressed in liver. Obvious species differences in the tissue distribution of AADAC were detected in jejunum and kidney. In contrast to human and mouse, AADAC mRNA was also expressed in rat testis. However, it was not detected in rat epididymis adipose tissue. The AADAC expression levels in rat tissues were much lower than those in human and mouse tissues. Trickett et al. (2001) predicted the transcription factors associated with the expression of AADAC by comparing the sequences in the 5′-flanking regions between the human and rodent AADAC genes. However, no information on the transcription factors causing different expression levels between human/mouse and rat was obtained. In addition, for rat, the AADAC expression levels in females were approximately 1.5- to 3.0-fold higher than those in males. However, a putative estrogen receptor response element was not found in the 2000 bases of the 5′-untranslated region of rat AADAC genes by computer-assisted homology research. Further study on the regulatory mechanism of AADAC transcription will be necessary.
To further analyze the AADAC protein level in tissue microsomes, we performed immunoblot analysis after SDS-PAGE using liver, jejunum, pulmonary, and renal microsomes of each species (Supplemental Fig. 2A). In human, AADAC protein was expressed in liver and jejunum but not in pulmonary and renal microsomes, which was coincident with the mRNA expression (Fig. 1). However, in rodents, the bands were detected in lung at approximately 45 kDa, which is the molecular mass of AADAC (Supplemental Fig. 2A), although the expression of AADAC mRNA was not observed in lung (Fig. 1). In addition, although rat kidney showed lower AADAC mRNA expression and enzyme activity than liver (Figs. 1 and 3), the band intensities in rat kidney and liver were similar. To examine the possibility that the antibody can detect some proteins other than AADAC in rodents, immunoblot analysis after native PAGE was performed (Supplemental Fig. 2B). However, RRM revealed a band with the same molecular mass as recombinant AADAC and RLM. This result suggested that AADAC protein in rat kidney is moderately expressed. Considering that the enzyme activity in RRM was much lower than that in RLM, any factor(s) may be involved in the AADAC enzyme activity in rat kidney. However, the possibility that the antibody can detect some proteins other than AADAC could not be fully denied. The monoclonal mouse anti-human AADAC antibody used in this study was produced by using human AADAC partial recombinant protein with 100 amino acid residues as the immunogen, and the homologies of the corresponding region with rat and mouse AADAC were 69 and 74%, respectively. Before making strict comparisons of expression levels of AADAC protein among tissues and species, we must develop specific antibodies for the various human, mouse, and rat enzymes.
Flutamide is mainly metabolized to 2-hydroxyflutamide, a pharmacologically active metabolite, by CYP1A2 and is also hydrolyzed to FLU-1 (Katchen and Buxbaum, 1975; Schulz et al., 1988). N-Hydroxyl FLU-1, which is a metabolite of FLU-1 by CYP3A4, has been suggested to be associated with hepatotoxicity (Goda et al., 2006). We found that AADAC is the principal enzyme responsible for flutamide hydrolysis in human (Watanabe et al., 2009). The present study found that rodent AADAC also catalyzed flutamide hydrolysis (Fig. 3A). The flutamide hydrolase activity in HLM followed the Michaelis-Menten equation, but activities in HJM and HRM followed the substrate inhibition equation at low flutamide concentrations (Fig. 3B; Supplemental Fig. 1). Our previous study investigated flutamide hydrolase activities in human tissue microsomes only at a high concentration (0.5 mM) (Watanabe et al., 2009). We will identify the enzyme catalyzing the flutamide hydrolysis at a low concentration in the near future. The kinetics for flutamide hydrolysis by recombinant rodent AADAC and rat tissue microsomes or MPM were fitted to a different equation (Fig. 3, A, C, and D). Thus, the contribution of other esterases to flutamide hydrolysis in rodent tissues was suggested. One of the candidate esterases involved in flutamide hydrolysis in rodents is CES, which is responsible for the biotransformation of a number of clinical drugs. Although rodents have multiple CES enzymes compared with humans (Holmes et al., 2010), the tissue distribution and functional characterization have not been fully elucidated. Further study about their distribution and substrate specificity will be necessary to clarify the contributions of AADAC and CES to flutamide hydrolysis.
Phenacetin is primarily metabolized to acetaminophen by CYP1A2 and is also hydrolyzed to p-phenetidine (Butler et al., 1989). p-Phenetidine is thought to be further metabolized to N-hydroxyphenetidine, the metabolite that possibly causes nephrotoxicity and hematotoxicity (Wirth et al., 1982; Jensen and Jollow, 1991). Our recent study found that AADAC is the principal enzyme responsible for the phenacetin hydrolysis in humans (Watanabe et al., 2010). The present study clarified that recombinant human and mouse AADAC showed higher catalytic efficiency in phenacetin hydrolysis than rat AADAC (Fig. 4; Table 3). In tissue microsomes, the CLint values of phenacetin hydrolysis reflected the expression levels of AADAC mRNA (Fig. 1; Table 3). This result suggested the high contribution of AADAC to phenacetin hydrolysis in the three species. It was reported that the mutagenicity of phenacetin in Salmonella typhimurium TA100 is detected only in the presence of liver S9 fractions from hamster but not in those from rat (Bartsch et al., 1980; Matsushima et al., 1980). Because the phenacetin hydrolase activity (phenacetin concentration: 1.4 mM) in hamster liver microsomes was 150-fold higher than that in RLM (Nohmi et al., 1983), it is probable that the high phenacetin hydrolase activity could induce the toxicity of phenacetin. However, there are no data about the sensitivity to phenacetin toxicity in these three species.
In flutamide and phenacetin hydrolase activities, the Km values in some tissue microsomes were different from those by recombinant AADAC (Tables 2 and 3). The difference in Km values between tissue microsomes and recombinant AADAC may be explained as follows: 1) the maximum substrate concentration was not sufficiently high to determine the Km values because of the limited solubilities of flutamide and phenacetin in the incubation mixture; 2) some factors expressing in tissue microsomes might alter the affinity of AADAC toward flutamide and phenacetin; 3) the activities in some tissue microsomes might be too low to calculate the appropriate Km values; and 4) especially in flutamide hydrolysis, other enzymes would be involved in the hydrolase activity.
Rifamycins such as rifampicin and rifabutin have the potential to induce various drug-metabolizing enzymes (Grange et al., 1994). In addition, rifampicin is suggested to be associated with hepatotoxicity (Gangadharam, 1986). Our recent study demonstrated that 25-deacetylrifamycins, the principal metabolites of rifamycins by AADAC in human, have lower induction potency and toxicity than rifamycins (Nakajima et al., 2011). Of interest, different from human AADAC, rodent AADAC could not catalyze rifampicin hydrolysis (Fig. 5A). Supporting this result, rifampicin hydrolase activities were detected in HLM and HJM in which AADAC was highly expressed but were scarcely detected in rodent tissue microsomes. It was reported that 25-deacetylrifampicin is detected in human plasma as the principal metabolite, whereas it is hardly detected in rat and rabbit plasma (Tenconi and Beretta, 1970; Chan, 1987). Binda et al. (1971) reported that dog tissues were also unable to hydrolyze rifampicin. Although we analyzed the hydrolysis of rifabutin (0.1 mM) and rifapentine (0.1 mM), which are rifamycin derivatives, their hydrolyzed metabolites were also not formed in MLM and RLM (data not shown). Thus, the 25-deacetylrifamycins might be a specific metabolite in human. The amino acid identities between human and rat or mouse AADAC are not high (68.2 and 69.9%, respectively). The differences between species in the relevant distances between sites of metabolism on substrates and the active serine residue in AADAC, the number of hydrogen bonds and π–π stacking interactions between substrates and the active site, and compound lipophilicity might partly explain the differences in the substrate recognition. Because an alternative metabolic pathway to hydrolysis in rifampicin metabolism has not been reported in rodents, it is conceivable that rifampicin is hardly metabolized in rat compared with human.
In conclusion, the present study clarified species differences in AADAC expression in tissues and enzyme activities. This is the first study that investigated AADAC species differences in drug metabolism. The substrate specificity of AADAC differs among species. The results obtained in this study will provide valuable information about species differences in drug pharmacokinetics and toxicity.
Authorship Contributions
Participated in research design: Kobayashi, Fukami, Nakajima, and Yokoi.
Conducted experiments: Kobayashi, Nakajima, and Watanabe.
Contributed new reagents or analytic tools: Kobayashi and Watanabe.
Performed data analysis: Kobayashi and Fukami.
Wrote or contributed to the writing of the manuscript: Kobayashi, Fukami, and Yokoi.
Acknowledgments
We acknowledge Brent Bell for reviewing the manuscript.
Footnotes
This study was supported by the Japan Society for the Promotion of Science [Grant-in-Aid for Young Scientists (B) 21790148].
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
↵
 The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.ABBREVIATIONS:
- CES
- carboxylesterase
- AADAC
- arylacetamide deacetylase
- FLU-1
- 4-nitro-3-(trifluoromethyl)phenylamine
- RT
- reverse transcription
- PCR
- polymerase chain reaction
- HLM
- human liver microsomes
- HJM
- human jejunum microsomes
- HRM
- human renal microsomes
- PAGE
- polyacrylamide gel electrophoresis
- RRM
- rat renal microsomes
- MJM
- mouse jejunum microsomes
- MRM
- mouse renal microsomes
- MLM
- mouse liver microsomes
- MPM
- mouse pulmonary microsomes.
- Received September 29, 2011.
- Accepted December 29, 2011.
- Copyright © 2012 by The American Society for Pharmacology and Experimental Therapeutics