Abstract
We quantified, by liquid chromatography tandem mass spectrometry, transporter protein expression of BSEP, MATE1, MRP3, MRP4, NTCP, and OCT1 in our human liver bank (n = 55) and determined the relationship between protein expression and sex, age and genotype. These data complement our previous work in the same liver bank where we quantified the protein expression of OATPs, BCRP, MDR1, and MRP2. In addition, we quantified and compared the interspecies differences in expression of the hepatobiliary transporters, corresponding to the above human transporters, in liver tissue and hepatocytes of male beagle dogs, cynomolgus monkeys, Sprague-Dawley rats, and Wistar rats. In all the species, the sinusoidal OATPs/Oatps were the most abundant hepatic transporters. However, there were notable interspecies differences in the relative abundance of the remaining transporters. For example, the next most abundant transporter in humans and monkeys was OCT1/Oct1, whereas it was Mrp2 and Ntcp in dogs/Wistar rats and Sprague-Dawley rats, respectively. In contrast, the protein expression of the efflux transporters BCRP/Bcrp, MDR1/Mdr1, MRP3/Mrp3, MRP4/Mrp4, and MATE1/Mate1 was much lower across all the species. For most transporters, the expression in the liver tissues was comparable to that in the unplated cryopreserved hepatocytes. These data on human liver transporter protein expression complete the picture of the expression of major human hepatobiliary transporters important in drug disposition and toxicity. In addition, the data on expression of the corresponding hepatobiliary transporters in preclinical species will be helpful in interpreting and extrapolating pharmacokinetic, pharmacological, and toxicological results from preclinical studies to humans.
Introduction
Hepatotoxicity is a major cause of failure in drug development (Kaplowitz, 2001). During drug development, the Food and Drug Administration requires in vivo repeated-dose toxicity studies of a drug candidate to be conducted in at least two animal species (one of which must be a nonrodent) before initiating human studies (http://www.fda.gov/downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm292340.pdf). Typically, these species are mouse or rat and dog or monkey. To interpret hepatotoxicity data generated in animals and predict their clinical relevance, it is important to understand the potential interspecies differences in mechanisms of toxicity of drugs. One such mechanism is transport of drugs into and out of the liver, which can result in interspecies differences in hepatic concentration of the drug (Lai, 2009). For example, a drug may rapidly be cleared from the liver in one animal species because of high expression of a hepatobiliary transporter. As a result, the hepatic concentrations of the drug will be low. In contrast, another species, which has much lower expression of the same transporter, may demonstrate elevated hepatic concentration of the drug, potentially causing hepatotoxicity. Therefore, it is important to compare the expression of transporters in liver tissue of humans and animals routinely used in toxicological studies. In addition, allometric scaling has been successfully used to predict human pharmacokinetics (PK) based on studies conducted in preclinical species (Huang and Riviere, 2014). However, these methods generally perform poorly for transporter substrates due to species differences in the substrate specificity, tissue distribution, and relative abundance of transporters (Chu et al., 2013). Quantitative knowledge of species differences in transporters, especially at the protein and functional level, will be useful to fill this knowledge gap and help improve allometric scaling of PK and drug-drug interactions from preclinical species to humans.
Thus far, interspecies differences in expression of transporters have mostly been compared by quantifying the expression of the mRNA of the transporter (by quantitative polymerase chain reaction) or protein (by Western blotting) in the tissues of species of interest. However, both methods have shortcomings. Often, for transporters, the mRNA expression does not correlate well with protein expression. For example, we and others have shown that mRNA expression of transporters is not always correlated with protein amounts in human livers (Ohtsuki et al., 2012; Prasad et al., 2013). Western blotting has the disadvantage that it is semiquantitative and, in the absence of pure protein standards, does not allow comparison of expression across proteins or species due to differences in affinity of the antibodies used. Transporter protein quantification, using surrogate signature peptides and liquid chromatography-mass spectrometry (LC-MS/MS), overcomes these limitations (Li et al., 2009a,b; Ito et al., 2011; Ohtsuki et al., 2012; Groer et al., 2013; Prasad et al., 2013, 2014). Briefly, enzymatic digestion (commonly by trypsin) is performed on target transporter protein and one or more of the liberated unique signature peptides is quantified by LC-MS/MS. The corresponding synthetic peptide as well as heavy labeled peptide is used as the calibrator and internal standard, respectively. This method allows simultaneous measurement of multiple transporters, and if there is sequence identity, the same signature peptide(s) can be used to quantify the same transporter protein in other species. Although, the expression of several transporters (MRP2/Mrp2, BCRP/Bcrp, NTCP/Ntcp, MDR1/Mdr1, BSEP/Bsep) in human and animal liver tissues and hepatocytes have been compared using this approach, this comparison did not include all relevant hepatic drug transporters or animal species studied here (Li et al., 2009b,c; Qiu et al., 2013).
In the present study, we quantified the protein concentration of BSEP, MATE1, MRP3, MRP4, NTCP, and OCT1 in human livers (n = 55). This extends our previous work in which we quantified several important human hepatic drug transporters, namely BCRP, MDR1, MRP2, and OATPs, in the same human liver bank (Deo et al., 2012; Prasad et al., 2013, 2014). The clinical relevance of these transporters has been summarized in recent reviews by the International Transporter Consortium (Giacomini et al., 2010; Hillgren et al., 2013). In addition we compared the expression of the above human hepatic transporters with those in animals, namely Bcrp, Bsep, Mdr1, Mrp2, Mrp3, Mrp4, Ntcp, Oatps, and Oct1 in liver tissues and unplated cryopreserved hepatocytes from male beagle dogs, cynomolgus monkeys, and Sprague-Dawley/Wistar rats.
Materials and Methods
Chemicals and Reagents.
Synthetic signature peptides for quantified transporters (Table 1) were obtained from New England Peptides (Boston, MA). The corresponding stable isotope-labeled (SIL) internal standards were purchased from Thermo Fisher Scientific (Rockford, IL). The ProteoExtract native membrane protein extraction kit was purchased from Calbiochem (Temecula, CA). Ammonium bicarbonate (98% purity) and sodium deoxycholate (98% purity) were obtained from Thermo Fisher Scientific and MP Biomedicals (Santa Ana, CA), respectively. BCA protein assay and the in-solution trypsin digestion kit were obtained from Pierce Biotechnology (Rockford, IL). High-performance liquid chromatography-grade acetonitrile (99.9% purity), methanol (99.9% purity) and formic acid (≥99.5% purity) were purchased from Fischer Scientific (Fair Lawn, NJ). Deionized water was generated from a Q-Gard 2 Purification Pack water purifying system (Millipore, Bedford, MA). Iodoacetamide and dithiothreitol were obtained from Pierce Biotechnology.
Multiple Reaction Monitoring parameters of peptides (calibrator and internal standard) selected for targeted analysis of hepatobiliary transporters in human (h), dog (d), monkey (m), and rat (r)
The labeled amino acid residue of the internal standard is shown in bold and italic.
Liver Samples and Hepatocytes.
Fifty-five liver tissue samples from the human liver bank of the School of Pharmacy, University of Washington, were used. Procurement (approved by the University of Washington Human Subjects Division), characteristics, and storage of these samples have been previously described (Paine et al., 1997), with additional details reported by Prasad et al. (2014). The subjects were Caucasian (n = 51), one Asian male, and three black males not of hispanic origin. Subject age range was 9 to 70 years and comprised of 29 women and 26 men. The majority of the livers were obtained from organ donors who met with accidental death (e.g., trauma from vehicular accidents, subarachnoid hemorrhage, cerebrovascular accident) and were harvested from breathing donors who were perfused with University of Wisconsin solution for organ transplant purposes. Six male beagle dog and six male Sprague-Dawley rat liver tissue samples were obtained from Bioreclamation IVT (Hicksville, NY). Ten male cynomolgus monkey liver tissue samples from Indonesian origin were provided by Merck (White House Station, NJ). Cryopreserved hepatocytes from three male beagle dogs and five male cynomolgus monkeys were purchased from Life Technologies (Carlsbad, CA). Cryopreserved hepatocytes from three Sprague-Dawley rats were obtained from Triangle Research Laboratories (Charlottesville, VA). Ten male liver tissue samples and one batch of cryopreserved hepatocytes from Wistar rats were provided by AstraZeneca (Boston, MA).
Peptide Selection.
Peptide sequences of the transporter proteins were obtained from two databases, Uniprot and PubMed. Transmembrane helices in transporter proteins were predicted using two different methods, TMHMM (Krogh et al., 2001) and Phobius (Kall et al., 2007). Whenever possible, two unique signature peptides, not present in any other known proteins, were selected for quantification of each transporter (Table 1) based on previously reported criteria (Kamiie et al., 2008; Prasad and Unadkat, 2014). Briefly, the selected peptides were between 9 and 16 amino acid residues, not embedded in the membrane or containing any known polymorphic variations or posttranslational modifications. In addition, peptides susceptible to degradation (e.g., containing methionine or cysteine residues) were not selected. Continuous R and K sequences (RR, RK, KR, and KK) were excluded to avoid miscleavages by trypsin. Whenever possible, we used the same signature peptides to quantify transporter protein expression in multiple species (see Table 1).
Sample Preparation.
Total membrane protein (in triplicate) was isolated from liver tissue or unplated cryopreserved hepatocytes as described previously (Prasad et al., 2014). Then, 100 µl of 2.0 mg/ml (or lower concentration) of liver tissue or hepatocytes membrane protein was incubated with 20 µl of dithiothreitol (100 mM) and 50 µl of ammonium bicarbonate buffer (50 mM, pH 7.8). After incubation at 95°C for 5 minutes, 20 µl of iodoacetamide (200 mM; an alkylating agent) were added to the mixture, followed by incubation at room temperature for 20 minutes in the dark. To concentrate the sample, ice-cold methanol (0.5 ml), chloroform (0.2 ml), and water (0.2 ml) were added to each sample. After centrifugation at 4°C for 5 minutes at 16,000 g, the pellet was washed once with ice-cold methanol (0.5 ml) and was resuspended with 40 µl of reconstitution solution [mixture of equal volume of 3% sodium deoxycholate (w/v) and 50 mM ammonium bicarbonate buffer]. Finally, the protein sample was digested with 20 µl of trypsin. Protein to trypsin ratio was 25:1 (w/w). After 24-hour incubation at 37°C, the digestion reaction was quenched by 30 µl of labeled peptide internal standard (SIL) cocktail (prepared in 50% acetonitrile in water containing 0.1% formic acid; final concentration of SIL: 0.1–0.25 nM). The samples were centrifuged at 5000 g for 5 minutes at 4°C, and 5 µl of the supernatant was introduced into the LC-MS/MS system.
The calibrators (in duplicate) were prepared by spiking peptide standards into the extraction buffer II of the membrane protein extraction kit. The quality control (QC) samples were prepared in triplicate by spiking peptides into extraction buffer II (three concentrations at low, medium, and high level of the calibration curve) or pooled liver membrane (two concentrations at medium and high level of the calibration curve).
Liquid Chromatography-Tandem Mass Spectroscopy Analyses.
Waters Xevo TQS tandem mass spectrometer coupled to Waters Acquity UPLC system (Waters, Hertfordshire, UK) operated in electrospray positive ionization mode was used for liquid chromatography-tandem mass spectroscopy (LC-MS/MS) analyses of the signature peptides. The mass spectrometry conditions were as follows: capillary 3.5 kV, source offset 30 V, source temperature 500°C (see Table 1 for analyte-specific parameters). The transitions from doubly charged parent ion to singly charged product ions for the analyte peptides and their respective SIL peptides were monitored (Table 1). The signature peptides and SIL peptide internal standards were separated on a Kinetex C18 column (100 mm × 3.0 mm i.d., 2.6 μm) with a Security Guard column (C18, 4 mm × 2.0 mm) from Phenomenex (Torrance, CA). Mobile phases (0.4 ml/min) consisted of 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B). The gradient program was: 0–3 minutes: 3% B; 3–6 minutes: 3–12.5% B; 6–8 minutes: 12.5–18% B; 8–13 minutes: 18–35% B; 13–15 minutes: 35–70% B; 15–16 minutes: 70–80% B; 16–17.1 minutes: 80–3% B; and 17.1–20 minutes: 3% B. Method validation was conducted on intraday accuracy and precision, as well as stability (freeze and thaw, bench-top, and autosampler conditions) as described previously (Prasad et al., 2013). The acceptable coefficient of variation was ≤15% and ≤ 5% for QC samples prepared in extraction buffer II and pooled liver membrane, respectively. The signal in the QC samples prepared in pooled liver membrane was corrected for the signal from the endogenous expression of the transporter.
mRNA Expression and Genotyping of Human Liver Tissue Samples.
The procedure for mRNA extraction and quantification, as well as genotyping of the human liver samples was previously reported (Naraharisetti et al., 2010; Prasad et al., 2013).
Statistical Analysis.
Data are reported as mean ± S.D. The Mann-Whitney rank order U test (α = 0.05) was used to compare protein expression between two groups.
Results
Analytical.
The calibration curves for the signature peptide were linear over two orders of magnitude (r2 greater than 0.995) and the lower limit of quantification (LLOQ) for all signature peptides were in the subfemtomole range (on column) (Table 1). The error and coefficient of variation of the QC samples was less than 25%, showing good accuracy and precision, respectively. Twenty-four-hour-trypsin digestion of the transport proteins was found to achieve maximum digestion, because no further increment in the yield of the peptide was observed beyond this time (data not shown). Where possible, we tested the recovery of the methanol-chloroform enrichment method versus the nonenrichment method by examining the final reported values of transporter protein expression obtained by both methods. The recovery of the methanol-chloroform enrichment method was on average ∼116% (range 87–130%). Thus no significant loss was observed during the enrichment process. Because purified transporter protein standards are currently not available, we assumed that the extraction efficiency for membrane protein from liver tissues or hepatocytes was complete and that the transporter protein was completely converted to its signature peptide(s) by trypsin digestion. All signature peptides were found to be stable during sample preparation (i.e., when exposed to three cycles of freeze and thaw, storage on bench-top for 6 hours, and in autosampler for 48 hours).
Although our goal was to use two signature peptides to measure the expression of each transporter protein, for some transporters only one peptide was detectable with acceptable MS sensitivity. Additionally, for four transporters, mMrp2, rMdr1, rOatp1a1, and rOatp1b2, significant differences (> 1.7-fold; P < 0.05) in protein quantification between the two signature peptides was observed. We attribute this to different degrees of trypsin digestion or the signal approaching LLOQ. Consequently, the results reported here were based on the peptides that generated the higher protein concentrations. Finally, we failed to find suitable signature peptides with desired MS sensitivity for quantification of dMate1, dNtcp, and mMate1.
Interindividual Variability in Transporter Expression in Human Livers.
In the narrative below and in data analyses, we included our previously published data on the expression of BCRP, MDR1 MRP2, OATP1B1, OATP1B3, and OATP2B1 in human liver to allow comparison of these data with those generated here (Deo et al., 2012; Prasad et al., 2013, 2014). These previous published data were generated from the same liver tissues and using a method identical to that used here.
In human livers, OATPs, OCT1, and NTCP comprised the most abundant sinusoidal influx transporters, whereas BSEP and MRP2 were the major canalicular efflux transporters (Fig. 1; Supplemental Table 1). Other canalicular efflux transporters (BCRP, MATE1, and MDR1) as well as sinusoidal efflux transporter (MRP3) showed lower expression (Fig. 1; Supplemental Table 1). MRP4 expression was below LLOQ. The interindividual variability in transporter protein expression, calculated as fold-range in transporter expression (maximum/minimum expression) was 2.8, 4.7, 3.7, 7.2, and 14 for BSEP, MATE1, MRP3, NTCP, and OCT1, respectively. Except for MATE1, which was significantly (P < 0.05) but weakly correlated with age (r2 = 0.16), the expression of the remaining transporters was independent of age. In addition, the expression of all the transporters was independent of sex.
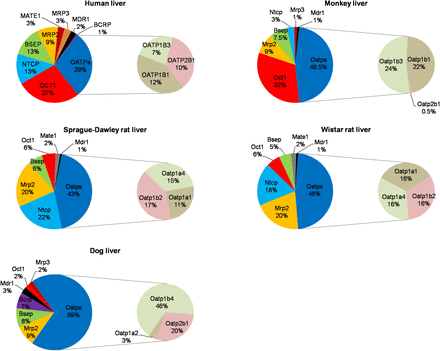
Relative abundance of quantifiable transporters in liver tissue of humans, beagle dogs, cynomolgus monkeys, Sprague-Dawley rats, and Wistar rats. Data are expressed as percent of total transporter protein expression in the liver tissue of the respective species. Expression data of human BCRP, MDR1, MRP2, and OATPs were obtained from our previous publications (Deo et al., 2012; Prasad et al., 2013, 2014). The expression of MRP4/Mrp4 was below LLOQ (0.6 fmol/µg membrane protein) in all the species studied.
Effect of Genotype on Human BSEP and OCT1 Protein Expression.
Some reported SNPs of BSEP and OCT1 were observed in the liver bank (Table 2). OCT1 was the only transporter where the protein expression was genotype dependent. Livers homozygous for the nonsynonymous SLC22A1 single nucleotide polymorphism (SNP), L160F, had 1.3-fold higher OCT1 protein (5.0±1.9 fmol/µg membrane protein) expression than the corresponding heterozygous livers (3.7±1.5 fmol/µg membrane protein) (P < 0.05). Subjects harboring other OCT1 or BSEP SNPs did not affect protein expression. No linkage disequilibrium between the various SNPs was observed for BSEP or OCT1.
Frequency of BSEP and OCT1 single nucleotide polymorphisms detected in the UW liver bank
Protein-Protein and mRNA-Protein Correlation of Transporters in Human Livers.
Protein-protein expression of the transporters was poorly correlated or nonexistent (r2 < 0.2; Supplemental Fig. 1). Likewise, the correlation between mRNA and protein expression was poor or nonexistent (r2 < 0.2; data not shown).
Comparison of Expression Profiles of Hepatobiliary Transporters in Human, Dog, Monkey, and Rat.
Relative abundance of transporter protein expression (i.e., percent of total transporter protein) in the investigated species (including humans) showed some similarity but also marked differences from that in human (Fig. 1). In all species, the OATPs/Oatps were the most abundant transporters. However, the next most abundant transporter differed. In humans and monkeys it was OCT1/Oct1, whereas it was Mrp2/Ntcp in rats and Mrp2 in dogs. Interestingly, Ntcp expression in the monkey ranked markedly lower than in humans, although its absolute expression was similar to that in humans (Fig. 2). Of the transporters studied, OATPs/Oatps, OCT1/Oct1, and NTCP/Ntcp, as well as MRP2/Mrp2 and BSEP/Bsep showed high relative expression in the sinusoidal and canalicular membrane, respectively (Fig. 1). When OATPs/Oatps expression was compared within each species, dOatp1b4 (i.e., dog Oatp1b4) and mOatp1b1/1b3 demonstrated the highest relative abundance, whereas the human OATP1B1, OATP1B3, and OATP2B1 and rat Oatp1a1, Oatp1a4, and Oatp1b2 showed comparable relative expression (Fig. 1). In addition, relatively high expression of NTCP/Ntcp was observed in human and rat (Fig. 1), whereas dNtcp was not quantified in dogs because we failed to find signature peptides with desired MS sensitivity for this transporter. In contrast, the protein level of the sinusoidal efflux transporters MRP3/Mrp3 and MRP4/Mrp4 was low across all the species tested. MRP3/Mrp3 expression was detectable only in human, dog, and monkey, whereas MRP4/Mrp4 expression was below LLOQ for all the species tested.
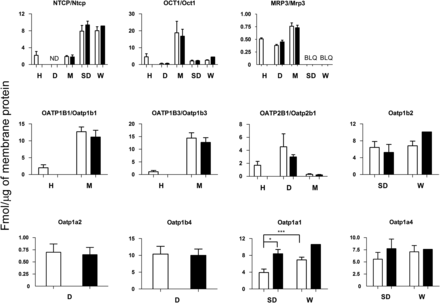
Expression profiles of sinusoidal transporters in liver tissue (open bars) and unplated cryopreserved hepatocytes (closed bars) of beagle dogs (D), cynomolgus monkeys (M), Sprague-Dawley (SD) rats, and Wistar (W) rats was compared with that in human (H) liver tissue (n = 55). Data on expression of human OATPs were obtained from Prasad et al. (2014). The expression of MRP4/Mrp4 was below LLOQ in all species. The sample size of liver tissue/hepatocytes was 6/3, 10/5, 6/3, 10/1 for dog, monkey, Sprague-Dawley rate, and Wistar rat, respectively. Only the difference in transporter expression between liver tissue and hepatocytes or between the rat strains was tested, and significant difference in expression is denoted by *P < 0.05 and ***P < 0.001 as determined by the Mann-Whitney rank order U test. BLQ, below LLOQ; ND, not determined.
The actual expression profiles of hepatobiliary transporters in liver tissue and hepatocytes from dog, monkey, and rat are shown in Figs. 2 and 3 and Supplemental Table 1. In the sinusoidal membrane, the rank order of the absolute OCT1/Oct1, NTCP/Ntcp, MRP3/Mrp3 expression level was monkey > human > rat > dog, rat > monkey ≈ human, and monkey > human ≈ dog > rat, respectively (Fig. 2; Supplemental Table 1). With respect to OATPs/Oatps, monkey showed markedly higher expression in Oatp1b1 and Oatp1b3 compared with those in human (OATP1B1 and OATP1B3), whereas dog showed highest expression of Oatp2b1, followed by human OATP2B1. Monkey showed the lowest expression of Oatp2b1 (Fig. 2; Supplemental Table 1). Among the canalicular transporters, MDR1/Mdr1 and BCRP/Bcrp had lower expression compared with MRP2/Mrp2 and BSEP/Bsep (Fig. 3; Supplemental Table 1). The rank order for MRP2/Mrp2 expression was rat > monkey > dog ≈ human. BCRP/Bcrp was quantifiable in only human and dog (dog > human; Fig. 3; Supplemental Table 1). The expression of MDR1/Mdr1 and BSEP/Bsep was similar in all the species investigated. Some differences were observed in Oatp1a1, Mate1, and Mdr1 protein expression between the two rat strains (Figs. 2 and 3; Supplemental Table 1). Except for rOatp1a1 (Sprague-Dawley rat), Mdr1 (dog and monkey), rMrp2 (Sprague-Dawley rat), the expression of hepatobiliary transporters in unplated cryopreserved hepatocytes was similar to that in liver tissue of the same species (Figs. 2 and 3; Supplemental Table 1). Protein-protein correlations of transporters in animal liver are shown in Supplemental Figs. 2 and 3. Although some of these correlations show high r2 values, these should be interpreted with caution due to the small number of liver samples studied.
Expression of canalicular transporters in liver tissue (open bars) and unplated cryopreserved hepatocytes (closed bars) of beagle dogs (D), cynomolgus monkeys (M), Sprague-Dawley (SD) rats, and Wistar (W) rats was compared with that in human (H) liver tissue (n = 55). Data on expression of human BCRP, MDR1, and MRP2 were obtained from our previous publications (Deo et al., 2012; Prasad et al., 2013). The sample size of liver tissue/hepatocytes was 6/3, 10/5, 6/3, 10/1 for dog, monkey, Sprague-Dawley rate, and Wistar rat, respectively. Only the difference in transporter expression between liver tissue and hepatocytes or between the rat strains was tested, and significant difference in expression is denoted by *P < 0.05, **P < 0.01, and ***P < 0.001 as determined by the Mann-Whitney rank order U test. BLQ, below LLOQ; ND, not determined.
Discussion
The University of Washington human liver bank provides a unique resource to investigate the interindividual variability in the protein levels of transporters. This work, together with our previous reports on the expression of a number of important drug transporters in this liver bank, provide a comprehensive picture of protein expression of clinically relevant sinusoidal and canalicular transporters in human livers (Deo et al., 2012; Prasad et al., 2013, 2014). Furthermore, we quantified a range of hepatic transporters in the liver tissues and hepatocytes of beagle dogs, cynomolgus monkeys, Sprague-Dawley rats, and Wistar rats and compared their expression with that in human liver tissues. These data will aid in interpretation and extrapolation of transporter-mediated hepatic elimination of drugs from animals to humans.
Except for OCT1, the variability in protein expression of the remaining transporters [including BCRP, MDR1, MRP2, MRP3, NTCP, and OATPs (Deo et al., 2012; Prasad et al., 2013, 2014)] was between 2.8- and 8-fold. No correlation was found between the expression of these transporters and sex, and MATE1 was the only transporter among those investigated that exhibited significant correlation with age. To better predict interindividual variability in hepatic disposition of a drug, it is important to determine the correlation in protein expression of the transporters (or metabolic enzymes or both). In the present study, poor or no correlation was observed in protein expression of the transporters (r2 < 0.2). In addition, as we observed before, poor or no correlation was observed between mRNA and protein expression of the transporters, indicating that mRNA expression does not reflect the protein expression of these transporters (Prasad et al., 2013).
OCT1 is involved in the hepatic uptake of metformin and plays an important role in the PK and pharmacodynamics of metformin. Previous studies demonstrated the influence of genetic variation in SLC22A1 on metformin pharmacological action. Reduced transport function of SLC22A1 allelic variants [e.g., rs12208357 (R61C), rs34130495 (G401S), and rs34059508 (G465R)] impairs hepatic uptake of metformin as demonstrated by decreased oral volume of distribution and glucose-lowering effects of the drug (Shu et al., 2007, 2008). For example, in vitro, compared with the reference allele, SLC22A1 SNPs, rs72552763 and rs12208357 showed decreased OCT1-mediated metformin uptake, whereas rs628031 and rs683369 did not affect metformin transport activity (Shu et al., 2003, 2007). In contrast, except for the minor increase in expression (1.3-fold) observed for SLC22A1 SNP rs683369, no significant change in OCT1 expression was observed for any of the investigated allelic variants (Table 2). Therefore, the reduced in vitro metformin transport observed for SLC22A1 SNPs rs72552763 and rs12208357 might be explained by factors such as elevated Km or decreased plasma membrane expression with no change in total membrane (including intracellular) expression of OCT1. Indeed, SLC22A1 SNP rs12208357 demonstrated reduced plasma membrane and enhanced cytosolic expression in comparison with SLC22A1 reference allele (Shu et al., 2007). Also, our expression data did not reflect the reduced Vmax of metformin transport observed for the SLC22A1 SNP rs72552763 (Shu et al., 2007). Perhaps this occurred because of reduced turnover number rather than protein expression of OCT1. In addition to OCT1, we also investigated the influence of BSEP SNPs on its expression. Neither of the two BSEP SNPs, rs2287622 and rs497692, showed altered BSEP expression, which is consistent with a previous study that the expression of BSEP reference gene (as measured by Western blotting) was not significantly different from its allelic valiant rs2287622 (V444A) (Meier et al., 2006).
Our data on hepatic transporter protein expression as well as the rank of expression among human and animals were similar to those published previously for BCRP/Bcrp, BSEP/Bsep, MRP2/Mrp2, and NTCP/Ntcp in human, beagle dog, rhesus/cynomolgus monkey, and Sprague-Dawley rat (Li et al., 2009b,c; Qiu et al., 2013). However, the present study greatly expanded the hepatobiliary transporters studied in preclinical species routinely used in drug development. In addition, our data now provide a complete picture of the interspecies differences in hepatobiliary transporter expression in humans and animals. Such information can be used to explain variation in transport activity in hepatocytes from these species. Ishizuka et al. (1999) showed that in vitro transport clearance across the bile canalicular membrane for 2,4-dinitrophenyl-S-glutathione (DNP-SG, a known Mrp2 substrate) in rat was eightfold higher than that in dog, whereas Km values for ATP-dependent uptake of DNP-SG into canalicular membrane vesicles were comparable between the two species, indicating that rat and dog Mrp2 have similar affinity (Km) for DNP-SG transport. Therefore, our observation of higher expression of Mrp2 in rat versus dog liver (four-fold) likely contributes to the difference in DNP-SG accumulation in rat canalicular membrane vesicles. Similarly, another study demonstrated that the rank order of hepatocyte elimination half-lives of the MRP2/Mrp2 substrates, glutathione-methylfluorescein and calcein, was human > monkey > dog > rat (Li et al., 2008). In addition, the BCRP/Bcrp-mediated pheophorbide efflux by human and dog hepatocytes was more efficient than that in monkey and rat, and MDR1/Mdr1-mediated 3,3-diethyloxacarbocyanine iodide efflux was minimal in hepatocytes of all origins. All these data are consistent with the rank order of expression of these transporters observed in our study. Although others have reported that the expression of hepatobiliary transporters in sandwich cultured rat and human hepatocytes varies with the culturing process (Li et al., 2009a), we found that the expression of hepatobiliary transporters in unplated cryopreserved hepatocytes was similar to that in liver tissue for most transporters and species.
MRP4/Mrp4 mediates hepatic basolateral efflux of drugs (e.g., rosuvastatin and ganciclovir) and serves as a compensatory route of excretion under cholestatic conditions (Gradhand et al., 2008; Pfeifer et al., 2013). In the present study, we were not able to measure MRP4/Mrp4 expression in liver tissue and hepatocytes from both human and animals likely because of the relatively high LLOQ of the signature peptides (shown in Table 1) and its low expression in liver tissues. Indeed, MRP4 protein expression in healthy human liver tissue has been measured by immunostaining, but this expression is much lower than that in livers from patients with cholestatic disease (Gradhand et al., 2008). In addition, we could quantify Bcrp only in liver tissue from dogs but not from monkeys and rats. As we used the same signature peptide to quantify Bcrp expression in all the species studied (same LLOQ), consistent with results obtained from a previous study (Li et al., 2009b), these data indicate that Bcrp expression was higher in dog liver compared with that in monkey or rat liver. However, this does not signify that Bcrp is not important in the disposition of drugs in the rat as this depends entirely on fraction of drug transported by Bcrp. If fraction of drug transported by hepatic Bcrp in the rat is large (e.g., 0.7), it will be the most important transporter in the biliary excretion of the drug.
There are some limitations to the present study. First, although it would be ideal to have purified transporter proteins as standards, this is currently not possible. Therefore, our method is based on absolute quantification of surrogate peptides. To translate such quantification to absolute transporter protein quantification, trypsin digestion must be complete. This may not be the case despite the fact that we optimized trypsin digestion. Second, we assumed that transporter protein extraction from the membrane is complete. Third, our expression data are for total cellular membrane and not purified plasma membrane. However, as we have shown, routine high-throughput isolation of the latter is not possible (Kumar et al., 2014). Therefore, we propose that if the expression of the transporter in plasma membrane versus that in total membrane is similar in liver tissue and in transfected cell lines (expressing the transporter of interest), in vitro-in vivo extrapolation and allometric scaling of transporter-mediated disposition of drugs are valid and should be possible. Finally, because of poor LC-MS sensitivity to certain signature peptides (e.g., Bcrp, MRP4/Mrp4), we failed to quantify the expression of a few transporters (e.g., Bcrp and MRP4/Mrp4). This limitation could potentially be overcome by improving the sensitivity of the assay.
In summary, the above data on human liver transporter protein completes the picture of the expression of clinically relevant hepatobiliary transporters in our liver bank and will facilitate future investigations on drug disposition and toxicity in the liver and interindividual variability in these measures. In addition, the above data on protein expression of the corresponding hepatobiliary transporters in preclinical species will now enable us to interpret and extrapolate PK, pharmacological, and toxicological results from preclinical species to humans.
Acknowledgments
Genotyping data were kindly provided by Dr. Yvonne Lin, Department of Pharmaceutics, University of Washington.
Authorship Contributions
Participated in research design: Wang, Prasad, Salphati, Chu, Gupta, Hop, Evers, Unadkat.
Conducted experiments: Wang, Prasad.
Contributed new reagents or analytic tools: Salphati, Chu, Gupta, Hop, Evers, Unadkat.
Performed data analysis: Wang.
Wrote or contributed to the writing of the manuscript: Wang, Prasad, Salphati, Chu, Gupta, Hop, Evers, Unadkat.
Footnotes
- Received October 16, 2014.
- Accepted December 22, 2014.
L.S., X.C., A.G., C.E.C.A.H., and R.E. contributed equally to the research.
This study was supported by the University of Washington Research Affiliate Program on Transporters (UWRAPT) sponsored by AstraZeneca, Genentech, and Merck & Co., Inc. (http://sop.washington.edu/uwrapt).
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
Abbreviations
- BCRP
- breast cancer resistance protein
- BSEP
- bile salt export pump
- DNP-SG
- 2,4-dinitrophenyl-S-glutathione
- Km
- Michaelis constant
- LC-MS/MS
- liquid chromatography tandem mass spectrometry
- LLOQ
- lower limit of quantification
- MDR1
- multidrug resistance
- MRP
- multidrug resistance-associated protein
- NTCP
- sodium-taurocholate cotransporting polypeptide
- OATP
- organic anion-transporting polypeptide
- OCT1
- organic cation transporter 1
- PK
- pharmacokinetic
- QC
- quality control
- SIL
- stable isotope-labeled
- SNP
- single nucleotide polymorphism
- Copyright © 2015 by The American Society for Pharmacology and Experimental Therapeutics