Abstract
Generating accurate in vitro intrinsic clearance data is an important aspect of predicting in vivo human clearance. Primary hepatocytes in suspension are routinely used to predict in vivo clearance; however, incubation times have typically been limited to 4–6 hours, which is not long enough to accurately evaluate the metabolic stability of slowly metabolized compounds. HepatoPac is a micropatterened hepatocyte-fibroblast coculture system that can be used for continuous incubations of up to 7 days. This study evaluated the ability of human HepatoPac to predict the in vivo clearance (CL) of 17 commercially available compounds with low to intermediate clearance (<12 ml/min/kg). In vitro half-life for disappearance of each compound was converted to hepatic clearance using the well stirred model, with and without correction for plasma protein binding. Hepatic CL, using three individual donors, was accurately predicted for 11 of 17 compounds (59%; predicted clearance within 2-fold of observed human in vivo clearance values). The accuracy of prediction increased to 82% (14 of 17 compounds) with an acceptance criterion defined as within 3-fold. When considering only low clearance compounds (<5 ml/min per kg), which represented 10 of the 17 compounds, the accuracy of prediction was 70% within 2-fold and 100% within 3-fold. In addition, the turnover of three slowly metabolized compounds (alprazolam, meloxicam, and tolbutamide) in HepatoPac was directly compared with turnover in suspended hepatocytes. The turnover of alprazolam and tolbutamide was approximately 2-fold greater using HepatoPac compared with suspended hepatocytes, which was roughly in line with the extrapolated values (correcting for the longer incubation time and lower cell number with HepatoPac). HepatoPac, but not suspended hepatocytes, demonstrated significant turnover of meloxicam. These results demonstrate the utility of HepatoPac for prediction of in vivo hepatic clearance, particularly with low clearance compounds.
Introduction
The development of drug candidates with acceptable pharmacokinetic (PK) parameters continues to be a challenge (Kola and Landis, 2004). Prospectively predicting PK parameters, using in vitro metabolism data, is a critical approach to mitigating the risk of advancing compounds with poor PK characteristics in the clinic. Metabolism by the liver is responsible for the clearance of approximately 70% of marketed drugs (Wienkers and Heath, 2005). The pharmaceutical industry has increasingly relied on the predictive power of in vitro models, such as liver subcellular fractions or hepatocyte preparations, to identify compounds with acceptable clearance characteristics (i.e., lower clearance compounds suitable for once-daily dosing) (Carlile et al., 1997; Iwatsubo et al., 1997; Brandon et al., 2003; Fagerholm, 2007). Hepatocyte suspensions reflect all metabolic pathways of the liver and this integrated metabolic capability potentially offers some clear advantages for hepatocytes as the model of choice for predicting hepatic clearance (Lau et al., 2002; Brown et al., 2007; Fagerholm, 2007). A tendency of hepatocytes to underpredict metabolic clearance compromises this position (Hallifax et al., 2010).
One of the key challenges in using suspended hepatocytes for predicting hepatic clearance is the loss of activity of drug metabolizing enzymes that restricts the typical incubation period to 4–6 hours (Elaut et al., 2006; Stringer et al., 2008). This limited incubation time prohibits the accurate determination of the intrinsic clearance (CLint) of compounds that are slowly metabolized. Consequently, an in vitro hepatocyte model that can survive (incubate) longer would offer the possibility of overcoming this restriction (Obach, 1999; Lau et al., 2002; Andersson et al., 2004; Riley et al., 2005; Brown et al., 2007).
HepatoPac is a micropatterned coculture of human hepatocytes, supported by mouse embryonic 3T3 fibroblasts, that has been engineered to possess extended cell viability (typically up to 6 weeks) with retention of in vivo–like hepatocyte functions in culture (e.g., albumin secretion, urea synthesis, and drug metabolizing activities) (Khetani and Bhatia, 2008; Wang et al., 2010). This in vitro hepatocyte culture system was shown to replicate in vivo metabolite profiles after incubations for 7 days, without a change in medium, which allowed ample time for the accumulation of detectable levels of metabolites that are slowly formed or derived from sequential metabolic pathways (Wang et al., 2010). In this study, HepatoPac was used to experimentally determine the metabolic turnover of 17 commercially available drugs that are low to moderately cleared in vivo (mainly by the liver). Hepatic clearances (CLh) were then calculated using the well stirred model and compared with published human in vivo nonrenal clearance (CLnonrenal) values to determine the accuracy of predictions. In addition, the depletion of three slowly metabolized compounds was followed in suspended hepatocytes and compared with HepatoPac cultures.
Materials and Methods
Human HepatoPac cultures were acquired from Hepregen Corporation (Medford, MA) and prepared from three lots (unpooled) of cryoplateable hepatocytes (donor designations: RTM, 1, and 3). Alprazolam, atazanavir, atomoxetine, diazepam, diclofenac sodium salt, flecainide, glimepiride, lidocaine, meloxicam, midazolam, prednisolone, riluzole, risperidone, theophylline, tolbutamide, tomoxetine, Trypan Blue, voriconazole, (±)warfarin, and William’s E media were purchased from Sigma-Aldrich (St. Louis, MO). A solution of penicillin (10,000 U/ml) and streptomycin (10 mg/ml), and GlutaMax (100X) were purchased from Gibco/Life Technologies (Grand Island, NY). A 20-donor customized pool of cryopreserved hepatocytes (LOT 000) was obtained from Celsis IVT (Baltimore, MD). Insulin, transferrin, and selenium (ITS+) premix cell culture supplement was acquired from BD Gentest (Franklin Lakes, NJ).
Cell Culture
HepatoPac.
Plateable cryopreserved primary human hepatocytes were purchased from commercial vendors permitted to sell products derived from human organs procured in the United States by federally designated organ procurement organizations. The donors consisted of three female Caucasian donors (donor 1, donor RTM, and donor 3) aged 19, 61, and 54 years of age, respectively. Cryopreserved hepatocytes were thawed at 37°C for 90–120 seconds followed by dilution with 50 ml prewarmed Hepregen-customized and proprietary hepatocyte culture medium (HCM) (Hepregen Corporation). The cell suspension was spun at 100 g for 10 minutes. The supernatant was discarded, cells were resuspended in HCM, and viability was assessed using Trypan Blue dye exclusion (viability was at least 85%). Liver-derived nonparenchymal cells, as judged by their size (approximately 10 μm in diameter) and morphology (nonpolygonal), were consistently found to be less than 1% in these preparations.
To create micropatterned cocultures in 96-well plates, a hepatocyte pattern was produced by seeding hepatocytes on collagen-patterned substrates (500 μm island diameter, 1200 μm center-to-center island spacing) that mediate selective cell adhesion (Khetani and Bhatia, 2008). The cells were washed with medium 4–6 hours later to remove unattached cells, leaving approximately 3,350 attached hepatocytes on 14 collagen-coated islands per well and incubated in HCM. The cell count per well was determined by microscopically counting cells on one island and multiplying this number by 14 islands in the well. Mouse embryonic fibroblasts (3T3-J2) were seeded 12–24 hours later to create cocultures. For cell maintenance, culture medium was replaced every 2 days (64 μl per well). To determine accurate cell counts, human micropatterned co-cultures were created in 96-well plates as described above. After 18 to 24 hours in culture, hepatocyte islands (without fibroblasts) were fixed and then stained using H&E (hematoxylin and eosin) dye. Images of the hepatocyte islands were captured using 20X magnification. Cells were counted using the ImageJ software. Average count per island were determined by counting ≥ 5 islands per well from ≥ 5 wells/plate from ≥3 independent seedings. Total counts were calculated by multiplying the average count by the number of islands. Coefficient of variation was calculated from the standard deviation of cell counts and found to be ±10%.
HepatoPac cultures were maintained in HCM with 10% serum for 7 days prior to treatment. Culture medium was changed every other day. HepatoPac cultures were washed with 100 µl serum-free Dulbecco’s modified Eagle’s medium immediately prior to the addition of test substances. Incubations with the test compounds were conducted in protein-free Dulbecco’s modified Eagle’s medium specially formulated for HepatoPac cultures (Hepregen Corporation). Test compounds were added to duplicate wells at a final concentration of 0.1 µM except for atomoxetine (0.5 µM). At various time points (up to 168 hours), 192 µl quench solution was added to terminate reactions. Quench solution contained acetonitrile/water (60:40, v/v), with 0.1% acetic acid and 0.1 µM internal standard (nevirapine or 1-naphthyl-β-d-glucuronide). The well surface was scraped with the end of a pipette tip and the mixture of cell debris and solution was triturated three times before depositing into corresponding wells on a separate 750-µl, 96-well plate. Incubations containing only mouse fibroblast cells served as controls.
Suspended Hepatocytes.
The 20-donor pool of cryopreserved human hepatocytes was purchased from Celsis In Vitro Technologies (Baltimore, MD). The pool consisted of hepatocytes from 11 male and 9 female donors. The average age of the donors was 48 years and the ethnic constitution was 5 Hispanics and 15 Caucasians. Cryopreserved hepatocytes were reconstituted in warm William’s E media containing 10% fetal bovine serum, 100 nM dexamethasone, 100 U penicillin, 0.1 mg streptomycin, and 1% ITS+ premix solution from BD Gentest. Viability of hepatocytes after reconstitution was 95%. Incubations in suspensions were carried out for 6 hours in William’s E complete medium containing 100 nM dexamethasone, 100 U penicillin, 0.1 mg streptomycin, and 1% ITS premix solution in an atmosphere of 5% CO2 and air, at 37°C. Test compounds were incubated with 50,000 hepatocytes in a 50-µl volume. Control incubations were carried out in culture medium only.
Liquid Chromatography/Mass Spectrometry Analysis
The amount of parent compound was measured using multiple reaction monitoring on an ABSciex Qtrap 4000 mass spectrometer (Foster City, CA). The mass spectrometer was coupled to Agilent 1200 serial binary pumps (Santa Clara, CA). The aqueous and organic mobile phase consisted of 95:5 (v/v) water/acetonitrile and 95:5 (v/v) acetonitrile/water, respectively. The mobile phase contained 0.1% acetic acid. Samples were eluted through a Phenomenex Kinetex C18 column (2.6 µm, 100 Å, 50 × 2.1 mm; Phenomenex Inc., Torrance, CA), a Phenomenex Synergi Polar-RP column (4 μm, 80 Å, 150 × 2.0 mm), or a Waters Atlantis dC18 column (5 μm, 100 Å, 100 × 2.1 mm; Waters Corporation, Milford, MA). The major cytochrome P450 (P450) isoforms responsible for the metabolism of test compounds and the multiple reaction monitoring transitions used for detection of each compound are listed in Table 1.
MRM transitions and major P450 isoforms responsible for metabolism of the 17 test compounds
Data Analysis and Clearance Calculations
The percentage of the remaining test compound during the incubation was calculated by dividing the mass spectrometry peak area ratio at a certain incubation time to that at time zero. The elimination rate constant (kel) was calculated from the absolute value of the slope derived from the natural logarithm of the percentage of drug remaining plotted against time. The depletion half-life of the test compound was calculated as in Eq. 1.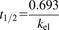 (1)Statistical significance in turnover of the parent compound was determined by comparing the slope of the linear regression line from the samples to that of the controls [compound incubated with medium (suspended hepatocytes) or fibroblasts (HepatoPac)] using an F test (GraphPad Prism, version 5.02; GraphPad Software, Inc., La Jolla, CA). The in vitro half-lives were used to calculate the intrinsic clearance (CLint) using scaling factors (Eq. 2). The parameters used for scaling are shown in Table 2 (Davies and Morris, 1993; Obach et al., 1997).
(1)Statistical significance in turnover of the parent compound was determined by comparing the slope of the linear regression line from the samples to that of the controls [compound incubated with medium (suspended hepatocytes) or fibroblasts (HepatoPac)] using an F test (GraphPad Prism, version 5.02; GraphPad Software, Inc., La Jolla, CA). The in vitro half-lives were used to calculate the intrinsic clearance (CLint) using scaling factors (Eq. 2). The parameters used for scaling are shown in Table 2 (Davies and Morris, 1993; Obach et al., 1997).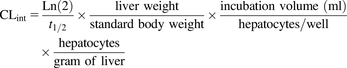 (2)
(2)
Scaling parameters to calculate CLint or CLh
The hepatic clearance (CLh) was calculated from CLint using the well stirred model (Eq. 3). CLh was calculated for each compound with or without correcting for plasma protein binding (fu) values that were obtained from the literature (fu = 1 when plasma protein binding was not considered). To compare in vivo CLint values with in vitro CLint values, the in vivo CLint was calculated by rearranging Eq. 3 into Eq. 4.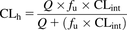 (3)Eq. 4:
(3)Eq. 4: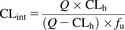 (4)
(4)
In vivo human nonrenal blood clearance values (CLnonrenal) for the 17 studied compounds were calculated by taking into account the total plasma clearance values (CL), blood to plasma partition ratio (Rb), and renal excretion of each compound (Eq. 5), where CL is the total plasma clearance and Frenal is the fraction of drug excreted by renal elimination. The plasma and blood clearance values for these compounds are summarized in Table 3. In this manuscript, CLnonrenal was assumed equal to CLh.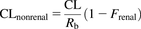 (5)
(5)
Human in vivo plasma and blood clearance values
The overall percentage turnover of alprazolam, tolbutamide, and meloxicam in HepatoPac and suspended hepatocytes was calculated from the in vitro–derived kel value using a one-phase elimination model (Eq. 6) instead of the percentage remaining at the final time point. This strategy ideally uses data from all of the time points to interpolate the value.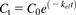 (6)
(6)
Monte Carlo Simulation
Monte Carlo simulations (MCSs) were conducted to model the relationship between percentage turnover and accuracy of CLint prediction. GraphPad Prism software (version 5.02) was used to conduct the simulations. Simulations were performed for 1, 5, 15, 30, 45, and 60% depletion of the parent compound using a one-phase elimination model with the plateau constrained to zero. The percentage depletion values were converted to elimination rate constants using Eq. 1. The elimination rate constants were subsequently used for the simulations. Gaussian relative variability was defined with a standard deviation of 15%. For each turnover value, 1000 simulations were conducted and the resulting half-lives were converted to CLint using Eq. 2 and the parameters from Table 3. The percentages of CLint values from each simulation that were within 2-fold of the true CLint value were plotted against the percentage of drug turnover. A hyperbolic equation (Eq. 7) was fitted to the data to obtain the percentage turnover value that was required to predict the corresponding CLint within 2- or 3-fold in 95% of cases. In Eq. 7, “A” is the percentage of experiments predicting within 2- or 3-fold of the actual CLint, D is the percentage of depletion of compound, and KD is the percentage of depletion of the compound when 50% of the experiments predict within 2- or 3-fold of the actual CLint (the values of slope factor h were 1.07 and 1.04 for within 2-fold and within 3-fold, respectively). (7)
(7)
Results
Accuracy of Clearance Prediction Using HepatoPac.
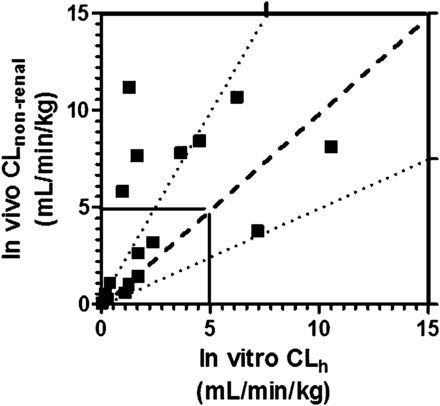
The predicted CLh of 10 compounds with low in vivo CLnonrenal in humans (<5 ml/min per kg) and 7 compounds with intermediate in vivo CLnonrenal (>5 and <15 ml/min per kg) are shown in Table 4. CLh values were calculated based on the parent depletion profiles in HepatoPac and were compared with the observed in vivo CLnonrenal values for the 17 compounds (Table 4). The comparison of mean predicted CLh values are plotted against the observed CLnonrenal values (Fig. 1). Using the mean CLh values for all three human donors and including correction for plasma protein binding, HepatoPac cultures predicted within 2-fold of the in vivo CLnonrenal for 11 of 17 compounds [65% accuracy; 7 of 10 compounds with low clearance (70%) and 4 of 7 compounds with intermediate clearance (57%)]. Using 3-fold as the criterion for accuracy of prediction, 14 of 17 compounds were predicted [82%; 10 of 10 low clearance compounds (100%) and 4 of 7 intermediate clearance compounds (57%)]. When plasma protein binding was not included in the well stirred model, the accuracy of prediction was much lower. Only 2 of 17 (12%) compounds were predicted within 2-fold of in vivo CLnonrenal [0 of 10 compounds with low clearance and 2 of 7 (29%) compounds with intermediate clearance], and 7 of 17 (41%) compounds were predicted within 3-fold of in vivo CLnonrenal [0 of 10 compounds with low clearance and 7 of 7 (100%) compounds with intermediate clearance].
Summary of CLh predictions for 17 compounds using HepatoPac incubations from three donors
Correlation between in vitro predicted CLh from HepatoPac and observed in vivo CLnonrenal for 17 compounds with low and intermediate clearance. The dashed line represents a perfect correlation. The dotted lines represent boundaries between a 2-fold underprediction and a 2-fold overprediction. The solid line marks the boundary between low and intermediate CLnonrenal values.
The accuracy of the prediction of CLh was similar between donors with only donor 3 showing lower accuracy at 2-fold (47%) compared with the other two donors at 83% and 71% (Table 4). None of the compounds showed greater than 10% turnover in fibroblast-only control plates (data not shown). Representative parent compound depletion graphs for compounds with varying in vitro CLint values (diazepam, theophylline, and atazanavir) are shown in Fig. 2 together with the fibroblast-only controls showing no turnover for up to 7 days.
Representative plots of parent compound depletion for theophylline (●), diazepam (▴), and atazanavir (▪) (each point represents the average of two replicates). The corresponding open symbols represent fibroblast-only controls (two replicates per experiment).
Comparison of Metabolic Turnover of Alprazolam, Meloxicam, and Tolbutamide Using HepatoPac and Suspended Hepatocytes.
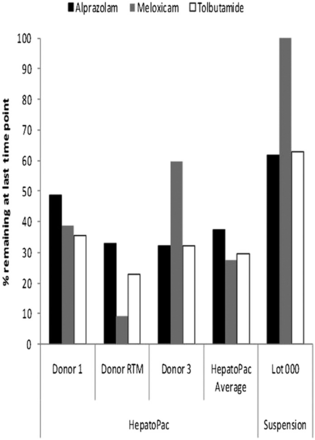
Incubations of HepatoPac cultures and suspended hepatocytes were compared for levels of turnover and clearance prediction. As expected, based on the larger number of hepatocytes in suspended cultures compared with HepatoPac, suspended hepatocytes generated a significantly greater percentage of turnover (not corrected for cell numbers) for alprazolam and tolbutamide. However, suspended hepatocytes did not show any turnover of meloxicam. Comparing the elimination rate constants using the full incubation times of 168 hours for HepatoPac and 6 hours for suspended hepatocytes, HepatoPac was found to provide 1.62- and 2.08-fold greater turnover (based on the average kel from each donor) for alprazolam and tolbutamide, respectively, in comparison with suspended hepatocytes (Fig. 3). These differences will be donor dependent and a better comparison would be provided by using the same donor in both systems.
Comparison of the percentage of parent compound remaining at the end of the incubation period in HepatoPac versus suspended hepatocyte incubations (alprazolam, meloxicam, tolbutamide).
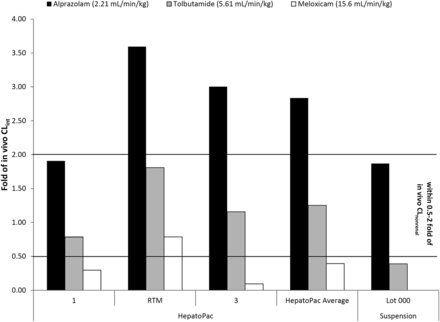
Appropriate scaling parameters were applied to determine intrinsic clearance and the well-stirred model was used to calculate the in vivo CLh for each compound. After averaging the values from each donor, HepatoPac predicted tolbutamide CLh to within 2-fold of the in vivo CLnonrenal and alprazolam and meloxicam CLh to within 3-fold of the in vivo CLnonrenal. The suspended hepatocyte incubations predicted CLh of alprazolam to within 2-fold and tolbutamide to within 3-fold of CLnonrenal. Since no turnover was observed for meloxicam with hepatocyte suspensions, CLh is expected to be under-predicted by greater than 3-fold in suspended hepatocytes (Fig. 4).
Comparison of the accuracy of prediction of CLnonrenal between HepatoPac and suspended hepatocytes.
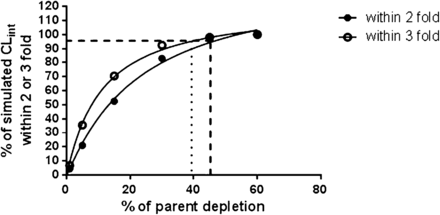
MCSs were conducted with the goal of determining the minimum clearance value (CLint) that both HepatoPac and suspended hepatocytes could predict. The simulation assumed bioanalytical variability of 15% of standard deviation (i.e., to be able to distinguish an extent of turnover from differences due to normal analytical variability). To achieve a success rate of 95% accurate prediction within 2- or 3-fold, the minimum percentages of parent depletion are 46.2 and 39.3%, respectively (Fig. 5). Using these values and the typical incubation conditions for HepatoPac and suspended hepatocytes as described in methods, the longest half-life that a compound can be considered to exhibit this extent of depletion is 191 or 233 hours in HepatoPac and 6.80 or 8.33 hours in suspended hepatocytes (within 2- and 3-fold of actual CLint, respectively). These half-life values correspond to CLint of 2.92 and 3.57 ml/min per kg in HepatoPac and 4.28 and 5.23 ml/min per kg in suspended hepatocytes (within 3- and 2-fold of actual CLint, respectively), indicating a 1.46-fold greater sensitivity for HepatoPac.
Universal relationship describing the percentage turnover of parent required to achieve an accurate prediction of CLint. The CLint values from 1000 simulations were obtained using the one-phase decay model and a 15% of standard deviation to represent bioanalytical or biologic variability. Turnover required to achieve results from 95% of experiments within 2-fold of the actual CLint is represented by a dashed line, and within 3-fold is represented by a dotted line.
Discussion
The ability to experimentally demonstrate turnover of slowly metabolized compounds in in vitro systems is dependent on enzyme concentration and incubation time. The lower limit of quantitation of CLh can vary depending on the analytical system being used. An alternative strategy that has been applied is to “bin” the CL prediction into categories of low, medium, or high clearance, which circumvents the challenges of accurately assessing the in vitro metabolic rates for low CL compounds (Wilkinson and Shand, 1975; Lavé et al., 1997). Although this strategy provides broad categories for clearance values, there are some limitations, particularly for compounds that are slowly metabolized. During lead optimization and identification, for chemical series exhibiting slow metabolism, it is still necessary to be able to differentiate compounds based on their predicted clearance. In addition, therapeutic dose predictions, based on clearance and target therapeutic concentrations, are valuable in anticipating drug supply and possible formulation needs for higher-dose, less soluble compounds, and in setting starting doses and dose escalation schemes for the first time in human studies.
In hepatocyte and microsomal incubations, CLint can be calculated from the formation of drug metabolites or disappearance of the drug. To calculate CLint from drug metabolite formation, authentic metabolite standards and well defined CL pathways are required. These requirements may not be practical in drug discovery in which the major enzymes responsible for drug CL may not have been elucidated or the authentic metabolites are not available to allow accurate quantitation of enzyme activity. Furthermore, compound-specific limitations, such as solubility, may not permit the determination of the required kinetic parameters, Vmax and Km. In early preclinical development, measuring the half-life of parent compound depletion is the easiest approach to determine in vitro metabolic CLint. In addition, by adopting multiple substrate concentrations, the requirement that substrate concentrations are well below their Km values, to be able to accurately determine CLint values, can be assessed (Obach, 2001). Both strategies can benefit from higher protein or cell concentrations to increase the metabolic rates of slowly metabolized drugs. However, higher protein concentrations can result in higher nonspecific binding that can affect substrate availability to drug metabolizing enzymes, ultimately leading to an underestimation of CLh (Obach, 2001). To circumvent the limited incubation time of microsomes, a relay method has shown promising results in which hepatocytes are replenished on multiple occasions to extend the overall incubation time (Di et al., 2012).
The loss of drug metabolizing enzyme function, which occurs after hepatocyte isolation and plating, reduces compound turnover and questions the ability of conventional long-term hepatocyte cultures to reflect in vivo metabolic capacity (Cross and Bayliss, 2000; Rodríguez-Antona et al., 2002). In the HepatoPac model, the heterotypic and homotypic interactions between human hepatocytes and mouse fibroblasts have been optimized through micropatterning in a multiwell plate format. This coculture has been shown to promote human hepatocyte morphology and phenotype, resulting in the maintenance of both viability and activity of hepatic drug metabolizing enzymes for an extended period with daily culture medium replenishment (Khetani and Bhatia, 2008). In our hands, P450 enzyme activity has been maintained consistently with human HepatoPac cultures, from different donors, typically for 2.5 weeks (in-house unpublished data). Furthermore, continuous drug incubations for up to 1 week in HepatoPac, without medium change, have been used to generate more complete, in vivo–like, metabolic profiles in comparison with conventional microsomal and hepatocyte incubations (Wang et al., 2010).
Using three low clearance drugs (alprazolam, meloxicam, and tolbutamide), a direct comparison of HepatoPac with human hepatocyte suspensions was conducted. From a theoretical perspective, focusing on the key differences between suspended hepatocytes and HepatoPac, in a 96-well format, there is a lower cell number per well for HepatoPac (3,350 hepatocytes/well versus 50,000 hepatocytes/well in hepatocyte suspensions). These differences correspond to a 15-fold lower source of drug metabolizing enzymes for HepatoPac. HepatoPac offers a longer incubation period (7 days for HepatoPac versus 6 hours for suspended hepatocytes), which translates to a 28-fold greater metabolic capacity. These comparisons obviously assume that all other factors between the two models are equivalent. It is important to note that a direct comparison of suspended hepatocytes and HepatoPac from the same human donor was not conducted due to the lack of availability of suspended hepatocytes from the same HepatoPac donors. Interdonor variability in drug metabolizing enzyme activity could also contribute to observed differences (see discussion below). In addition, the role of transporters in the clearance of drugs is becoming increasingly apparent (Shitara et al., 2013) and drug transporter expression and proper localization on hepatic membranes have been shown to differ between hepatocyte cultures and hepatocyte suspensions. For example, efflux transporters are internalized in suspensions (Bow et al., 2008) and uptake transporters have been reported to be preferentially depleted in sandwich-cultured hepatocytes (Tchaparian et al., 2011).
With the aforementioned caveats and assumptions in mind, considering only the differences in cell number and the incubation period, HepatoPac is expected to have 1.5-fold higher metabolic capacity than suspended hepatocytes. Based on MCS, the lowest scaled CLint values that could be accurately determined with HepatoPac are 2.92 ml/min per kg and 4.28 ml/min per kg for suspended hepatocytes. This difference in metabolic activity (1.46-fold) between HepatoPac and hepatocyte suspensions is roughly in line with the observed 1.62- and 2.08-fold higher turnover observed with alprazolam and tolbutamide, respectively, using HepatoPac compared with suspended hepatocytes (based on the average turnover using hepatocytes from three donors for HepatoPac; Fig. 3). In addition, meloxicam, which exhibited the slowest rate of depletion in HepatoPac, showed no perceptible depletion using suspended hepatocytes. Interestingly, the calculated CLnonrenal of meloxicam (0.089 ml/min per kg) is lower than that of alprazolam (0.61 ml/min per kg) or tolbutamide (0.31 ml/min per kg), but the calculated CLint for meloxicam (15.6 ml/min per kg) was faster than alprazolam (2.21 ml/min per kg) and tolbutamide (9.45 ml/min per kg). This discrepancy is rooted in the well stirred model (Eq. 3), which predicts that the high plasma protein binding of meloxicam is a major factor determining its CLnonrenal. As such, accurate CLint should be obtained in in vitro incubations that do not contain plasma proteins. However, the underprediction of CLint was still observed for meloxicam. The slow turnover of meloxicam in in vitro hepatocyte cultures such as hepatocyte suspensions or HepatoPac may also be limited by drug binding in the incubation. Drug binding to incubation components such as culture ware, fibroblasts, or hepatocytes was not evaluated in this study. Clearly, possible differences in specific P450 content between donors could also contribute to these differences. In comparison, Stringer et al. (2008) noted that perceptible turnover could only be observed with 3 of 24 reference drugs (13%) with low to intermediate in vivo CLint (ranging from 1 to 10 mL/min/kg).
The accuracy of prediction for low clearance compounds (10 drugs evaluated), when including correction for plasma protein binding, was 100% within 3-fold of in vivo CLnonrenal and 70% within 2-fold. Although this is a limited data set, the results are encouraging in that HepatoPac allows for sufficient turnover of compounds for measurement of intrinsic clearance. In contrast, the effect of plasma protein binding on the accuracy of CLnonrenal prediction for the intermediately cleared drugs was less clear as the difference in the accuracy of predictions with and without plasma protein binding was less extreme. A decrease in the percentage of compounds predicting CLnonrenal to within 2-fold (57 to 29%) was observed when ignoring plasma protein binding (i.e., setting the fraction of drug unbound to plasma protein to 1). However, when considering accuracy to within 3-fold, there was a marked improvement in accuracy from 57 to 100%.
The effect of plasma protein binding on the predictivity for intermediate clearance compounds requires a more extensive analysis, which is in progress. That said, a tendency to underpredict the clearance of intermediate to high clearance compounds when incorporating plasma protein binding, agrees with previous studies using conventional hepatocyte suspensions. A recently published PhRMA study made a recommendation to set fu=1 when using hepatocytes. In this study the majority of compounds (6 of 9) were intermediate to high clearance (Ring et al., 2011). In a retrospective analysis of in vitro studies using hepatocytes to predict the in vivo CL of 89 compounds, a bias to more greatly underpredict the intrinsic clearance of rapidly metabolized compounds was described using suspended hepatocytes (Hallifax et al., 2010). In addition, the authors found that the accuracy of prediction was affected by drug binding in the blood (including plasma proteins) such that accuracy improved when blood binding was low. The bias to more accurately predict the CLh of high clearance compounds when plasma protein binding is not accounted for is consistent with the trend observed in this study whereby for the intermediate clearance compounds, the exclusion of plasma protein binding does not adversely affect the overall accuracy of prediction. Several recent mathematical models that incorporate factors such as plasma albumin to liver albumin ratio and drug ionization may improve the ability to predict hepatocyte incubations, including HepatoPac, by reducing average prediction bias, but may not entirely eliminate it (Berezhkovskiy, 2011; Hallifax and Houston, 2012; Poulin et al., 2012).
The variability in calculated clearance values between hepatocytes from different donors indicates that donor-dependent variation in drug metabolizing activity (Shimada et al., 1994) is retained in plated or suspended hepatocytes (Ponsoda et al., 2001). Differences in predicted clearance values were observed for the three donors in this study, which reiterates one of the challenges with using hepatocytes for prediction of human clearance, that of interindividual variability in drug metabolizing enzymes. To mitigate these differences, suspended hepatocytes can be pooled, with the goal of generating an “average” human, similar to combining human liver microsomes from different donors. However, differences in plating efficiencies add a complexity to this process for HepatoPac and other plated hepatocyte cultures. An obvious contributor to interindividual variability is the polymorphic expression of P450s (e.g., with drugs metabolized by CYP2C9, CYP2C19, and/or CYP2D6). In this study, there was a trend for donor RTM to clear CYP2C9 substrates, diclofenac, glimepiride, and meloxicam, faster than donor 1 or donor 3. Clearance predictions using hepatocytes should characterize human donors and select, where possible, hepatocyte lots reflecting average enzyme activity for the major P450 isoforms or, when the contributing drug metabolizing enzyme(s) are known, correct CL for differences in enzyme activity, which can potentially be done by relative activity factors (Venkatakrishnan et al., 2000). Drug metabolizing enzyme polymorphisms or donor-dependent differences in response to culture conditions (Waring et al., 2003; Chen et al., 2011) have not been evaluated in this analysis but are being considered in future studies.
In summary, the extended drug incubation period with HepatoPac allowed for detectable metabolic turnover of low CL drugs. As a result, a high accuracy of prediction was achieved with low CL drugs. The ability of HepatoPac to accurately predict intermediate and high CL drugs is being investigated. The extended functionality of hepatocytes in HepatoPac suggests that additional important factors such as drug transporters, which can play a crucial role in drug clearance (Shitara et al., 2006), may also be appropriately represented in vitro. This is also a topic of further investigation.
Correction
Chan, Yu, Moore, Khetani and Tweedie, Meeting the Challenge of Predicting Hepatic Clearance of Compounds Slowly Metabolized by Cytochrome P450 Using a Novel Hepatocyte Model, HepatoPac
In the above article [Chan TS, Yu H, Moore A, Khetani SR, and Tweedie D (2013) Drug Metab Dispos 41: 2024-2032, https://doi.org/10.1124/dmd.113.053397], subsequent to publication of the article, the authors were made aware of an error in the number of cells per well which affects calculation of clearance parameters. In recalculating these parameters, the overall conclusions of the study have not changed. The text, Table 4, and Figure 4 have been revised to reflect the changes in the clearance values. Due to the number of locations in the article where the results needed to be updated to represent the corrected parameters, this corrected version is being published. A correction to correct spelling of author Salman R. Khetani was published in January 2014. The HTML and PDF versions of the article have been updated.
Authorship Contributions
Participated in research design: Chan, Yu, Moore, Khetani, Tweedie.
Conducted experiments: Chan, Yu, Moore.
Contributed new reagents or analytic tools: Chan, Yu, Moore.
Performed data analysis: Chan, Yu.
Wrote or contributed to the writing of the manuscript: Chan, Yu, Khetani, Tweedie.
Footnotes
- Received June 20, 2013.
- Accepted August 15, 2013.
S.R.K. is an equity holder in Hepregen Corporation.
T.S.C. and H.Y. contributed equally to this work.
Abbreviations
- C0
- initial concentration
- Ct
- concentration at a specified time during the incubation
- CL
- clearance
- CLh
- predicted hepatic clearance
- CLint
- intrinsic clearance
- CLnonrenal
- observed human in vivo nonrenal clearance
- fu
- fraction of drug unbound to plasma protein
- HCM
- hepatocyte culture medium
- ITS
- insulin, transferrin, and selenium
- kel
- elimination rate constant
- MCS
- Monte Carlo simulation
- P450
- cytochrome P450
- PK
- pharmacokinetic
- Q
- hepatic blood flow
- Rb
- red blood cell partitioning
- t1/2
- half-life
- Copyright © 2018 by The American Society for Pharmacology and Experimental Therapeutics