Abstract
In contrast to a single human carboxylesterase 2 (CES2) isozyme (hCE2), three CES2 genes have been identified in cynomolgus monkeys: mfCES2A, mfCES2B, and mfCES2C. Although mfCES2A protein is expressed in several organs, mfCES2B is a pseudogene and the phenotype of the mfCES2C gene has not yet been clarified in tissues. In previous studies, we detected an unidentified esterase in the region of CES2 mobility upon nondenaturing PAGE analysis of monkey intestinal microsomes, which showed immunoreactivity for anti-mfCES2A antibody. The aim of the present study was to identify this unidentified esterase from monkey small intestine. The esterase was separated on nondenaturing PAGE gel and digested in-gel with trypsin. The amino acid sequences of fragmented peptides were analyzed by tandem mass spectrometry. The unidentified esterase was shown to be identical to mfCES2C (XP_015298642.1, predicted from the genome sequence data). mfCES2C consists of 559 amino acid residues and shows approximately 90% homology with mfCES2A (561 amino acid residues). In contrast to the ubiquitous expression of mfCES2A, mfCES2C is only expressed in the small intestine, kidney, and skin. The hydrolytic properties of recombinant mfCES2C, expressed in HEK293 cells, with respect to p-nitrophenyl derivatives, 4-methylumbelliferyl acetate, and irinotecan were similar to those of recombinant mfCES2A. However, mfCES2C showed a hydrolase activity for O-n-valeryl propranolol higher than mfCES2A. It is concluded that the previously unidentified monkey intestinal CES2 is mfCES2C, which shows different hydrolytic properties to mfCES2A, depending on the substrate.
SIGNIFICANCE STATEMENT In the present research, we determined that mfCES2C, a novel monkey CES2 isozyme, is expressed in the small intestine and kidney of the cynomolgus monkey. Interestingly, mfCES2C showed a relatively wide substrate specificity for ester-containing compounds. These findings may, in early stages of drug development, support the use of in vitro–to–in vivo extrapolation for the intestinal hydrolysis of ester drugs in the cynomolgus monkey.
Introduction
Carboxylesterases (CESs; EC 3.1.1.1), members of the α,β-hydrolase fold superfamily, are important metabolic enzymes that catalyze the hydrolysis of various environmental toxicants, xenobiotics, and pharmaceutical agents. Compounds containing ester, amides, or thioesters can be hydrolyzed by CESs to carboxylic acids and their respective alcohols, amines, and thiols (Satoh and Hosokawa, 2006). Carboxylic acid esters designed as prodrugs, for example, irinotecan (CPT-11), capecitabine, temocapril, and oseltamivir, are also mainly metabolized by CESs, generating the active metabolites that show the pharmacological efficacy (Imai, 2006; Laizure et al., 2013).
CES1 and CES2 families are largely responsible for metabolism of drugs and prodrugs. In humans, there is a single isozyme in each of these families, that is, hCE1 is the CES1 isozyme and hCE2 the CES2 isozyme. Interestingly, hepatic and intestinal hydrolase activities are mainly dependent on the activities of hCE1 and hCE2, respectively (Hosokawa, 2008; Imai and Ohura, 2010). The different substrate specificities of hCE1 and hCE2 can be used in designing prodrugs (Imai et al., 2006).
Cynomolgus macaques are frequently used in drug metabolism studies owing to their physiologic and genetic similarities to humans. However, several isozymes of CESs are present in cynomolgus macaques. For example, in the CES1 family in cynomolgus macaques, two CES1 isozymes (mfCES1A and mfCES1B) have been detected as mRNA in several tissues (Uno et al., 2014). mfCES1A, the major CES1 isozyme, showed similar tissue distribution and hydrolase activity to hCE1 (Williams et al., 2011), and mfCES1B was also present in some organs.
On the other hand, three CES2 genes (mfCES2A, mfCES2B, and mfCES2C) have been reported (Williams et al., 2011; Uno et al., 2014), with expression profiles different from hCE2. It has been reported that the mRNA of mfCES2A is present in most organs (Uno et al., 2014), and the expression of mfCES2A protein has been confirmed in liver and intestine (Hosokawa et al., 1990; Igawa et al., 2016). The amino acid sequence of mfCES2A is 89% homologous with hCE2 (Supplemental Table 1). However, the hydrolytic preferences of mfCES2A recombinant protein for several substrates is different from that of hCE2 (Igawa et al., 2016). In contrast, mfCES2B gene is considered a pseudogene (Uno et al., 2019). We have also found a new transcript variant of mfCES2A, mfCES2Av2, in which exon 3 of mfCES2Av1, a wild type of mfCES2A, replaces exon 3 of mfCES2B. The mfCES2Av2 variant is 98% homologous with mfCES2Av1, and we have confirmed the expression of mfCES2Av1 and mfCES2Av2 proteins in cynomolgus monkey liver (Uno et al., 2019). The hydrolytic properties of mfCES2Av2 recombinant protein for several substrates were very similar to those of mfCES2Av1 (Uno et al., 2019). In contrast to mfCES2A, mfCES2C mRNA is specifically expressed in jejunum and kidney (Uno et al., 2014). However, there are few literature reports on the tissue expression of a mature mfCES2C protein or its substrate specificity. In our previous studies, nondenaturing PAGE of cynomolgus intestinal microsomes showed two major bands of proteins with esterase activity in the migration region of the CES2 family of enzymes (Taketani et al., 2007). The protein in the upper band was designated as the imfCES2A (Igawa et al., 2016), therefore it is assumed that the protein on the lower band is mfCES2C.
In this study, to identify and characterize the mfCES2C protein from small intestinal microsomes of cynomolgus monkeys, we separated the lower band protein on nondenaturing PAGE gel; the amino acid sequences of peptides obtained by in-gel tryptic digestion of the protein were analyzed by tandem mass spectrometry and the hydrolytic activities of mfCES2C were compared with those of mfCES2Av1 using their recombinant proteins expressed in human embryonic kidney (HEK) 293 cells.
Materials and Methods
Tissue Samples and Reagents.
Pooled liver and individual small intestinal microsomes (phenylmethylsulfonyl fluoride-free) of cynomolgus macaques (male, 4–6 years old, 3–6 kg) were kindly provided by Sekisui Medical Co., Ltd. (Ibaraki, Japan) and BoZo Research Center Inc. (Tokyo, Japan). Rabbit anti-mfCES2Av1 polyclonal antibodies were kindly provided by Dr. M. Hosokawa (Chiba Institute of Science, Chiba, Japan). Fluorophosphonate (FP)-biotin was synthesized as described by Liu et al. (1999). All other chemicals and reagents were of analytical grade and purchased from standard commercial suppliers. Animal experimental protocols were approved by the Ethics Review Committee for Animal Experimentation of Kumamoto University.
Nondenaturing PAGE.
The details of nondenaturing PAGE analysis were described in our previous report (Ohura et al., 2010). Microsomes from cynomolgus monkey liver and small intestine, and cytosol fractions of HEK293 cells expressing recombinant CES2 isozymes were placed on 7.5% nondenaturing polyacrylamide gel. After electrophoresis, the bands containing esterases were stained by complex formation between Fast Red TR hemi(zinc chloride) salt, a dyeing agent, and α-naphthol produced by hydrolysis of α-naphthyl acetate as a substrate of esterase.
Immunoblotting.
Immunoblotting was performed as described previously (Igawa et al., 2016). Briefly, microsomes from cynomolgus monkey liver and small intestine and cytosol fractions of HEK293 cells expressing recombinant CES2 isozymes were added to 7.5% polyacrylamide gel with SDS. After electrophoresis, proteins were transferred to a polyvinylidene difluoride (PVDF) membrane. After blockade with 5% skimmed milk in phosphate-buffered saline, the proteins were probed with rabbit anti-mfCES2Av1 polyclonal antibody. The membranes were incubated with peroxidase-conjugated rabbit IgG, and immunoreactive proteins were detected by Clarity Western ECL Substrate (Bio-Rad Laboratories, Inc., Berkeley, CA) using the Image Quant LAS 4000 (GE Healthcare Life Sciences, Piscataway, NJ).
Extraction of Peptides Digested by Trypsin in Nondenaturing Polyacrylamide Gel.
Nondenaturing PAGE was performed using two individual intestinal samples (No. 2 and No. 7). As shown in Fig. 1B and Supplemental Fig. 1, sample No. 2 showed two bands in the migration region of CES2 isozymes, whereas sample No. 7 showed only one band. The gels were cut out at the same positions as the respective bands of mfCES2Av1 and the unidentified esterase. Proteins contained in each gel piece were reduced with 10 mM dithiothreitol followed by alkylation with 55 mM iodoacetamide, and digested with Sequencing Grade Modified Trypsin (Promega Corporation, Madison, WI). Digested peptides were extracted in 50% acetonitrile including 5% formic acid and desalted by Stage-Tips (Rappsilber et al., 2003). The samples were resolved with 5% formic acid and analyzed by liquid chromatography–tandem MS (LC-MS/MS).
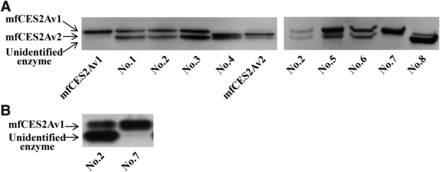
Expression of mfCES2 isozymes in cynomolgus monkey intestine. (A) Immunoblotting analysis using anti-mfCES2Av1 antibody. The proteins of small intestinal microsomes (3 μg) from eight cynomolgus monkeys and recombinant mfCES2A isozymes (3 μg) were subjected to SDS-PAGE and transferred electrophoretically to a PVDF membrane for immunostaining. (B) Esterase activity staining after nondenaturing-PAGE using α-naphthylacetate. Each lane contained intestinal microsomes (100 μg) from an individual cynomolgus monkey.
LC-MS/MS Analysis.
All LC-MS/MS analyses were performed on a Orbitrap Elite coupled to the EASY-nLC 1000 system (Thermo Fisher Scientific Inc., Waltham, MA). The electrospray ionization tip column packed in-house with ReproSil-Pur 120 C18-AQ (2.4 μm, 100 μm i.d. × 300 mm length; Dr. Maisch HPLC GmbH, Ammerbuch-Entringen, Germany) was used at a flow rate of 300 nl/min and the column oven maintained at 50°C. The peptides were separated by a linear gradient condition from 5% to 28% of acetonitrile in 0.125% formic acid for 100 minutes. Orbitrap Full MS scan spectra were collected in the range of m/z 350–2000 at a resolution of 12,000, followed by sequential ion trap collision-induced dissociation, and MS/MS spectra of the 20 most intense ions were obtained. Their spectra were analyzed by Core, a software tool suite for proteomics developed in the Gygi laboratory from Harvard Medical School (Huttlin et al., 2010), using Mascot (Matrix Science Ltd., London, UK) as a search engine and RefSeq macaca fascicularis sequence database supplemented with protein sequences from cRAP, a database of common contaminating proteins by the Global Proteome Machine Organization (112 sequences from http://www.thegpm.org/crap/index.html) and in-house registered sequences [e.g., CES2v3 (AIM58905.1)], using the following parameters: maximum missed cleavage: 2; static modification: carbamidomethylcysteine; variable modifications: methionine, serine, and threonine; mass tolerances for precursor and fragment ions: 50 ppm and 0.8 Da, respectively. Peptide- and protein-level false discovery rates were filtered to 1% using the target-decoy strategy (Elias and Gygi, 2007) to distinguish between correct and incorrect identifications.
Stable Expression of Recombinant CES2 Isozymes in HEK293 Cells.
Recombinant mfCES2Av1 and mfCES2C are stably expressed in HEK293 cells, as described previously (Igawa et al., 2016). Briefly, mfCES2Av1 and mfCES2C cDNA inserted into pTargeT vector (Promega Corporation) were transfected to HEK293 cells (American Type Culture Collection, Rockville, MD; passage 10–12), followed by selection of clones with resistance to G418. The confluent cells of stable clones were then homogenized in SET buffer (292 mM sucrose, 1 mM EDTA, and 50 mM Tris), and S9 fractions were separated as the supernatant by centrifugation (9000g, 20 minutes, 4°C). Cytosolic fractions were prepared as the supernatant after ultracentrifugation (105,000g, 1 hour, 4°C) of S9 fractions, owing to the presence of high amounts of recombinant CES isozymes in the cytosol, resulting from saturation of the KDEL receptor, which interacts with the C-terminal endoplasmic reticulum retention peptides of recombinant CES isozymes. The cytosolic fraction was stored at −80°C until use.
Determination of Expression Level of Recombinant mfCES2 in the Cytosol Fraction of HEK293 Cells.
The content of the recombinant mfCES2 isozymes in the cytosolic fraction was determined with FP-biotin using a previously described method (Abdel-Daim et al., 2018). Briefly, the recombinant mfCES2 isozymes in the cytosolic fraction and purified porcine liver esterase as a standard protein (Sigma-Aldrich Co. LLC, St. Louis, MO) were reacted with FP-biotin and subjected to SDS-PAGE. After electrophoresis, the proteins were transferred to a PVDF membrane, and proteins labeled with FP-biotin were detected by avidin-peroxidase with ImmunoStar LD (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan). Protein bands were quantified using Image J, a free software for densitometric analysis. The content of recombinant mfCES2 isozymes in the cytosolic fraction was calculated using the calibration curve of purified porcine liver esterase. The contents of mfCES2Av1 and mfCES2C in the cytosolic fractions were 0.60 and 0.15 nmol enzyme per mg cytosolic protein, respectively.
Measurement of In Vitro Hydrolytic Activity.
The cytosolic fractions of HEK293 cells expressing recombinant mfCES2 proteins were diluted to 5–100 μg/ml with 50 mM 2-[4-(2-hydroxyethyl)-1-piperazinyl] ethanesulfonic acid (HEPES) buffer, pH 7.4. After preincubation at 37°C for 5 minutes, each hydrolytic reaction was initiated by the addition of substrates (300 μM p-nitrophenyl derivatives, 300 μM 4-methylumbelliferyl acetate, 25 μM O-n-valeryl propranolol, and 50 μM irinotecan) dissolved in dimethyl sulfoxide (DMSO). For the inhibition experiment, loperamide, a specific inhibitor of hCE2, dissolved in DMSO, was added to the enzyme solution and preincubated at 37°C for 5 minutes. The reaction was started by adding O-n-valeryl propranolol. The final concentration of DMSO in the reaction mixture was less than 1%, which had no effect on hydrolytic activity.
The hydrolysis rate of p-nitrophenyl derivatives was spectrophotometrically determined by the concentration of p-nitrophenol at 405 nm. The concentration of propranolol, a hydrolysate of O-n-valeryl propranolol, was determined by high-performance liquid chromatography (HPLC), according to the method reported in our previous studies (Igawa et al., 2016). For the hydrolysis of 4-methylumbelliferyl acetate and irinotecan, the reaction was terminated by adding an equal volume of ice-cold acetonitrile. After centrifugation of the reaction mixture at 1600g for 10 minutes, H3PO4 (final concentrations 50 and 30 mM, respectively) was added to the supernatant, and their solutions analyzed by HPLC. The HPLC system comprised a pump (PU-980; JASCO), autosampler (AS-950; JASCO), fluorescence detector (FP-1520S; JASCO), column oven (CO-965; JASCO), and data application apparatus (ChromNAV chromatography data system; Jasco International Co. Ltd., Tokyo, Japan). The temperature of the column was maintained at 40°C. For the determination of 4-methylumbelliferone concentration, L-column3 C18 (5 μm, 4.6 mm i.d. × 150 mm length; Chemicals Evaluation and Research Institute, Tokyo, Japan) was used with a mobile phase of acetonitrile/water, 35:65 (v/v) at a flow rate of 1.0 ml/min. 4-Methylumbelliferone was detected at excitation and emission wavelengths of 323 and 477 nm, respectively. For the determination of SN-38 concentration, LiChrospher 100 RP-8 end-capped HPLC column (5 μm, 4.0 mm i.d. × 125 mm length; Merck KGaA, Darmstadt, Germany) was used with a mobile phase of acetonitrile/12 mM ammonium acetate (pH 4.0), 25:75 (v/v) at a flow rate of 1.0 ml/min. SN-38 was detected at excitation and emission wavelengths of 375 and 567 nm, respectively. Substrate and metabolite were clearly separated and were measured in a quantitatively linear range.
The initial hydrolytic activity was measured under reaction conditions in which less than 20% of substrate was hydrolyzed. Hydrolytic activity was expressed as the mole of hydrolysate per minute per millimole of recombinant CES enzyme. The activities of recombinant samples were calculated by subtraction of the activities of cytosolic fractions derived from mock-transfected HEK293 cells.
Data Analysis.
The hydrolytic activity data are represented as the mean ± S.D. of triplicate samples. Statistical analysis was performed by one-way analysis of variance, followed by Scheffe’s F test, and P-values < 0.05 were considered to be statistically significant.
Results
Interindividual Differences in mfCES2 Expression in the Small Intestine of Cynomolgus Monkeys.
Expression of mfCES2 isozymes was investigated using intestinal samples from eight monkeys. Figure 1A shows the result of immunoblotting analysis using anti-mfCES2Av1 antibody. Immunoreactive bands corresponding to mfCES2A were observed as bands representing either mfCES2Av1 or mfCES2Av2 in all intestinal samples. The wild type of mfCES2A, mfCES2Av1, was present in six intestinal samples, whereas samples No. 4 and No. 8 expressed mfCES2Av2. The band representing the previously unidentified enzyme was observed in seven intestinal samples, but not No. 7. Interestingly, the No. 7 sample showed only one band for mfCES2Av1. We selected sample No. 2 as a standard phenotype in its expression of both mfCES2 isozymes, and then compared the expression of mfCES2 isozymes in samples No. 2 and No. 7 by nondenaturing PAGE analysis. As can be seen in Fig. 1B, sample No. 2 showed two bands with weak and strong band intensities (slow and fast mobilities, respectively). Only one band, related to mfCES2Av1, was detected in sample No. 7.
Identification of Previously Unidentified Enzyme by LC-MS/MS Analysis.
After nondenaturing PAGE, gel pieces corresponding to the upper and lower bands of samples No. 2 and No. 7 were cut out as shown in Supplemental Fig. 1. The amino acid sequences of peptides generated by digestion of the proteins in each gel piece were determined by LC-MS/MS analysis. XP_005592244.2 (PREDICTED: cocaine esterase-like [Macaca fascicularis]) and XP_015298642.1 (PREDICTED: LOW QUALITY PROTEIN: cocaine esterase-like [Macaca fascicularis]) were estimated for the upper and lower bands, respectively, by an NCBI RefSeq Macaca fascicularis database search of peptide sequences. The numbers of peptide fragments matched for XP_005592244.2 in the upper bands of samples No. 2 and No. 7 were greater than those in their lower bands. The most peptide fragments matched for XP_015298642.1 were detected in the lower band of No. 2.
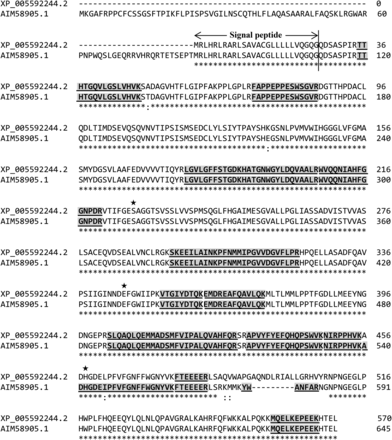
Figure 2 shows the amino acid sequences of XP_005592244.2 with the common peptide fragments observed in the gel piece of the upper bands of samples No. 2 and No. 7. These amino acid sequences were also compared with the mfCES2Av1 (GenBank accession number: AIM58905.1) reported by Uno et al. (2014). The amino acid sequence of XP_005592244.2 showed a high homology (96%) with AIM58905.1. The peptide fragments from the gel piece of the upper band were the best match with AIM58905.1.
Alignment of amino acid sequences estimated as a protein present in the upper band of intestinal samples No. 2 and No. 7 by LC-MS/MS analysis. The amino acid sequence of XP_005592244.2 (Gene Symbol: LOC102119346), estimated as a protein present in the upper band of both intestinal samples No. 2 and No. 7, was aligned with the amino acid sequence of mfCES2Av1 (GenBank No.: AIM58905.1) from Uno et al. (2014). Gray highlight with underline shows the amino acid sequences of peptides as determined by LC-MS/MS analysis. Star symbols show the amino acid residues of the catalytic triad.
Figure 3 shows the amino acid sequences of XP_015298642.1. The homology of the amino acid sequence of mfCES2C was 98% with XP_015298642.1. All peptide fragments from the gel piece of the lower band could be matched with the amino acid sequences of both XP_015298642.1 and mfCES2C (cCES2). Therefore, the esterases included in the upper and lower bands of sample No. 2 were predicted to be mfCES2Av1 and mfCES2C, respectively.
Alignment of amino acid sequences estimated as a protein present in the lower band of intestinal sample No. 2 by LC-MS/MS analysis. The amino acid sequence of XP_015298642.1 (Gene Symbol: LOC102120721), estimated as a protein present in the lower band of intestinal sample No. 2, was aligned with the amino acid sequence of mfCES2C (cCES2) from Williams et al. (2010). Gray highlight with underline shows the amino acid sequences of peptides as determined by LC-MS/MS analysis. Star symbols show the amino acid residues of the catalytic triad.
Preparation of Recombinant mfCES2Av1 and mfCES2C and Confirmation of the Phenotype of mfCES2C in Nondenaturing PAGE and Immunoblotting Analyses.
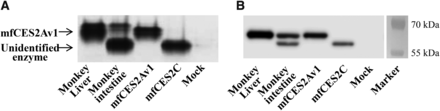
The recombinant mfCES2 isozymes were overexpressed in HEK293 cells and analyzed by nondenaturing PAGE and immunoblotting methods. The resulting isozymes were compared with those from cynomolgus monkey intestinal and hepatic microsomes to confirm the phenotype of the mfCES2Av1 and mfCES2C proteins. As shown in Fig. 4, both nondenaturing PAGE and immunoblotting analysis showed that the mobility of mfCES2Av1 recombinant protein was the same as the upper band in monkey liver and intestine. In contrast, mfCES2C recombinant protein was detected at the same mobility as the lower band of the cynomolgus monkey intestine, but it was not detected in the liver.
Expression of recombinant mfCES2 isozymes in HEK293 cells. (A) Esterase activity staining after nondenaturing PAGE using α-naphthylacetate. The protein samples (cynomolgus monkey liver microsomes, 10 μg; cynomolgus monkey jejunum microsomes, 50 μg; mfCES2Av1 cytosol fraction, 10 μg; mfCES2C cytosol fraction, 20 μg; mock-transfected cytosol fraction, 20 μg) were loaded in separate lanes. Arrows indicate bands corresponding to each CES isozyme. (B) Immunoblotting analysis using anti-mfCES2Av1 antibody. The sample proteins (cynomolgus monkey liver microsomes, 1 μg; cynomolgus monkey jejunum microsomes, 30 μg; mfCES2Av1 cytosol fraction, 1 μg; mfCES2C cytosol fraction, 2 μg; mock-transfected cytosol fraction, 2 μg) were subjected to SDS-PAGE and transferred electrophoretically to a PVDF membrane for immunostaining.
Hydrolytic Activities of Recombinant mfCES2Av1 and mfCES2C Proteins.
The hydrolase activities of recombinant mfCES2Av1 and mfCES2C were compared using several substrates: p-nitrophenyl derivatives, 4-methylumbelliferyl acetate, O-n-valeryl propranolol, and irinotecan. The results are shown in Table 1. All substrates tested were hydrolyzed by both mfCES2C and mfCES2Av1. The hydrolase activities of mfCES2C for p-nitrophenyl derivatives, 4-methylumbelliferyl acetate, and irinotecan were nearly the same as those of mfCES2Av1. However, mfCES2C hydrolyzed O-n-valeryl propranolol more rapidly than mfCES2Av1; the hydrolysis rates for the R-isomer and S-isomer with mfCES2C were 3.0- and 6.4-fold faster, respectively, than with mfCES2Av1. The preferential hydrolysis of the R-isomer of O-n-valeryl propranolol was more clear-cut for mfCES2Av1; the R/S ratio of hydrolase activity was 4.3 and 9.4 for mfCES2C and mfCES2Av1, respectively.
Comparison of hydrolase activities of mfCES2Av1 and mfCES2C
The cytosol fractions from HEK293 cells expressing recombinant CES2 isozyme were diluted with 50 mM HEPES buffer (pH 7.4) at 5–100 μg/ml. Substrates (p-nitrophenyl derivatives and 4-methylumbelliferyl acetate: 300 μM; O-n-valeryl propranolol: 25 μM; irinotecan: 50 μM) were incubated with the diluted cytosol fractions. Values represent the mean ± S.D. (n = 3).
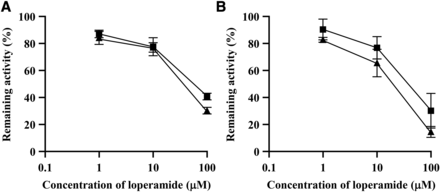
The inhibitory effect of loperamide, a selective hCE2 inhibitor, on the hydrolysis of O-n-valeryl propranolol was compared using recombinant mfCES2Av1 and mfCES2C. As shown in Fig. 5, the inhibitory effect of loperamide on the hydrolysis of O-n-valeryl propranolol was similar for mfCES2C and mfCES2Av1; approximately 80% of the hydrolase activities of mfCES2Av1 and mfCES2C for O-n-valeryl propranolol remained at 10 μM loperamide, whereas 60%–80% of activity was inhibited at 100 μM loperamide.
Inhibitory effect of loperamide on the hydrolysis of O-n-valeryl propranolol by recombinant mfCES2 isozymes. Hydrolysis activity for the R-isomer (A) and S-isomer (B) of O-n-valeryl propranolol (25 μM) by recombinant mfCES2 isozymes was measured in the absence (control) or presence of 1–100 μM loperamide. Control was set as 100%. Square and triangular symbols indicate mfCES2Av1 and mfCES2C, respectively. Each symbol represents the mean ± S.D. (n = 3).
Discussion
Cynomolgus monkeys express two CES2 isozymes, mfCES2A and mfCES2C. The mfCES2C mRNA is expressed specifically in the jejunum and kidney, in contrast to the ubiquitous expression of mfCES2A mRNA (Uno et al., 2014, 2019). Therefore, mature proteins of both mfCES2A and mfCES2C would be expected to be present in the small intestine of cynomolgus monkey. However, no evidence for the presence of mfCES2C proteins in the intestine has been presented until now. As shown in Fig. 1A, mfCES2A was detected as a slower migrating band on immunoblot analysis, and all individuals expressed either mfCES2Av1 or mfCES2Av2. The other, faster migrating band was detected in all samples except for No. 7, as shown in Fig. 1A. Two bands were clearly visible in the migration region of CES2 isozyme on nondenaturing PAGE analysis of sample No. 2 (Fig. 1B). Two bands had also been detected in our earlier nondenaturing PAGE analysis using intestinal microsomes from other monkeys (Taketani et al., 2007). These results suggest that the previously unidentified esterase of monkey intestine may be mfCES2C, which has an isoelectric point below that of mfCES2A, which is 4.7 (Hosokawa et al., 1990).
Fortunately, sample No. 7 was from a monkey who lacked the band representing the unidentified esterase. When the proteins separated on the nondenaturing PAGE gel were digested by the in-gel tryptic method, and the amino acid sequences of the peptides produced were analyzed, we could demonstrate, for the first time, that the fast-migrating band in the nondenaturing PAGE and immunoblot analysis was mfCES2C.
It is difficult to explain why mfCES2C migrates faster than mfCES2Av1 in immunoblot analysis. mfCES2C consists of 559 amino acid residues, two amino acids fewer than mfCES2Av1 (Supplemental Fig. 3); mfCES2C possesses two N-glycosylation sites at Asn111 and Asn276, in comparison with the single N-glycosylation site (Asn111) of mfCES2Av1; the sequence of mfCES2C differs from that of mfCES2Av1 by 43 amino acids. Thus, structural factors cannot explain the mobility shifts of the mfCES2 isozymes. A previous study (Uno et al., 2019) has shown that mfCES2Av2, differing from mfCES2Av1 only by 10 amino acid residues, still migrated faster than mfCES2Av1 in immunoblot analysis (shown in Fig. 1A). It is possible that differences in their protein structures after denaturation by SDS and/or the structure of their sugar chains may be responsible for these differences in mobility.
We also confirmed the presence of mfCES2C proteins in cynomolgus monkey kidney, as well as small intestine, on nondenaturing PAGE analysis. As shown in Supplemental Fig. 2, two bands relating to mfCES2Av1 and mfCES2C were detected at similar intensities in kidney microsomes. In a previous report, we also detected the mfCES2C band on nondenaturing PAGE analysis of the S9 fractions of cynomolgus monkey skin, but its intensity was lower than that of mfCES2A (Imai et al., 2019). In contrast to these organs, cynomolgus monkey liver possesses only mfCES2A. The expression profile of mfCES2C (intestine, kidney, and skin) was similar to that of human CES2 isozyme (hCE2) (Zhang et al., 2002; Wu et al., 2003). These data suggested that cynomolgus monkey has two CES2 isozymes with different expression profiles and that mfCES2C is the major isozyme in the intestine.
In this study, the hydrolytic ability of mfCES2C was compared with mfCES2Av1 using four different substrates. The residues of the catalytic triads (mfCES2Av1, Ser228-Glu346-His458; mfCES2C, Ser228-Glu345-His457), the four cysteines involved in the formation of disulfide bonds (mfCES2Av1 and mfCES2C, Cys95-Cys123 and Cys280-Cys291), and the endoplasmic reticulum retention signal of the C-terminal (His-Thr-Glu-Leu-COOH) are highly conserved in both mfCES2Av1 and mfCES2C. The residues of the oxyanion hole of mfCES2Av1 are Gly149 and Gly150, and those of mfCES2C are Gly149 and Ala150. Owing to the similar structures of these enzymes, the p-nitrophenyl derivatives, 4-methylumbelliferyl acetate, and irinotecan were likewise hydrolyzed by both mfCES2 isozymes (Table 1). However, the hydrolysis rate of O-n-valeryl propranolol by mfCES2C was faster than that by mfCES2Av1. In addition, the R/S ratio, an index of R-isomer-preferential hydrolysis, was decreased by 50% in mfCES2C (R/S ratio 4.3) in comparison with mfCES2Av1 (R/S ratio 9.4). We have previously reported that hCE2 hydrolyzes R- and S-isomers of propranolol derivatives equally and at a faster hydrolysis rate than did mfCES2Av1 (Igawa et al., 2016). We have also reported a docking simulation study of enzyme-propranolol derivatives, in which we showed that the enantioselectivity of mfCES2Av1 is caused by an amino acid residue change of Leu107 to Gln107 and the insertion of Met309, compared with hCE2 (Igawa et al., 2016). The amino acid residue at position 107 in mfCES2C is tyrosine, and Met309 is absent (Supplemental Fig. 3). These amino acid sequences are closer to hCE2 than mfCES2Av1. Furthermore, the amino acid residues of the oxyanion hole (Gly149 and Ala150) and two N-glycosylation sites (Asn111 and Asn276) are highly conserved between hCE2 and mfCES2C. It is therefore considered that the weaker enantioselectivity and faster hydrolysis rate of O-n-valeryl propranolol in mfCES2C compared with mfCES2Av1 might be related to changes in the amino acid residues around the active center and the number of N-glycosylation sites.
The inhibitory effect of loperamide, a potent selective inhibitor of hCE2, on the hydrolysis of O-n-valeryl propranolol was similar for both mfCES2Av1 and mfCES2C. Williams et al. (2011) reported that the IC50 value for the inhibition of hydrolysis of 4-methylumbelliferyl butyrate by loperamide in mfCES2C is 34.4 μM, which is almost the same as the result obtained in our hydrolysis inhibition experiment using O-n-valeryl propranolol as a substrate, as shown in Fig. 5. As loperamide shows similar inhibitory effects with respect to different substrates for both mfCES2Av1 and mfCES2C isozymes, loperamide may be an effective inhibitor for not only hCE2 but also mfCES2 isozymes. On the other hand, the IC50 value (0.562 μM) of the inhibitory effect of loperamide on the hydrolysis of 4-methylumbelliferyl butyrate by hCE2 is lower than that of mfCES2C (Williams et al., 2011). The inhibitory effect of loperamide on the hydrolysis of O-n-valeryl propranolol by hCE2 was also stronger than that by mfCES2 isozymes (Fig. 5; Supplemental Fig. 4). These results indicate that the inhibiting affinity of loperamide for mfCES2 isozymes is less than that for hCE2, suggesting slightly different binding mechanisms for hCE2 and mfCES2 isozymes.
In the present study, we identified the expression of mfCES2C as a protein in the small intestine and kidney of cynomolgus monkey. The amino acid residues near the active site in mfCES2C (oxyanion hole, deletion of Met309 and Tyr107) are more similar to those in hCE2 than in mfCES2Av1. We demonstrated that mfCES2C recognized the same substrates as mfCES2Av1. However, the exact hydrolytic functions of mfCES2A and mfCES2C and their contribution to the first-pass hydrolysis of prodrugs in the small intestine of cynomolgus monkey has not yet been fully clarified. Further investigations of their substrate specificity and expression levels in the small intestine will be required to simulate precisely in vitro and in vivo intestinal absorption and hydrolysis of ester drugs in the cynomolgus monkey and, thus, to demonstrate the suitability of these monkeys for use in the preclinical stages of drug development.
Acknowledgments
The authors thank Sekisui Medical Co., Ltd. (Ibaraki, Japan) and BoZo Research Center Inc. (Tokyo, Japan) for the kind gift of liver and intestinal microsomes of cynomolgus macaques, M. Hosokawa (Chiba Institute of Science) for his kind gift of anti-mfCES2Av1 antibody, and Tatsuya Inoue (Daiichi Sankyo RD Novare Co., Ltd.) for database maintenance and protein annotation.
Authorship Contributions
Participated in research design: Ohura, Igawa, Imai.
Conducted experiments: Ohura, Igawa, Tanaka, Matsumoto, Kasahara, Wada, Kubota, Uno.
Contributed new reagents or analytic tools: Ohura, Igawa, Imai.
Performed data analysis: Ohura, Igawa.
Wrote or contributed to the writing of the manuscript: Ohura, Igawa, Imai.
Footnotes
- Received August 8, 2019.
- Accepted December 2, 2019.
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
↵
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
Abbreviations
- CES
- carboxylesterase
- FP
- fluorophosphonate
- hCE1
- human carboxylesterase 1
- hCE2
- human carboxylesterase 2
- HEK
- human embryonic kidney
- HPLC
- high-performance liquid chromatography
- LC-MS/MS
- liquid chromatography–tandem mass spectrometry
- PVDF
- polyvinylidene difluoride
- Copyright © 2020 by The American Society for Pharmacology and Experimental Therapeutics