Abstract
The influence of microsomal concentration on unbound fraction (fumic), half-life (t1/2), apparent intrinsic clearance (CLint,app) and apparent Michaelis-Menten constant (Km,app) was examined for two compounds, one representative of high nonspecific binding to microsomes (compound A) and one representative of low (compound B). Kinetic parameters were estimated for the two probe compounds at two human microsomal protein concentrations (0.46 and 2.3 mg/ml) and cytochrome P450 concentrations (0.20 and 1.0 μM), representing a 5-fold difference in microsomal concentration. For compound A, fumic and CLint,app were inversely proportional to microsomal concentration. Conversely, theKm,app of compound A was proportional to microsomal concentration and the half-life was unchanged. For compound B, half-life was inversely proportional to microsomal concentration. In this case, fumic, CLint,app, andKm,app were not proportionally influenced. The experimental observations were entirely consistent with that predicted by a mathematical relationship between microsomal concentration, fumic, t1/2, CLint,app, and Km,app. These results demonstrate that when nonspecific binding is extensive, CLint,app is dependent on the arbitrary choice of microsomal concentration included in the incubation.
In recent years, several methods have been proposed to predict the human clearance of drug substances from data obtained in liver microsomes (Lave et al., 1997, 1999; Obach, 1997,1999; Obach et al., 1997). Because of their predictive utility and ease of use, high-throughput liver microsomal lability assays are widely employed in drug discovery to prospectively identify compounds that will have desirable pharmacokinetics in humans (Lave et al., 1999;Obach, 1999; Kohl and Steinkellner, 2000). The fact that nonspecific microsomal binding has the potential to confound the accurate prediction of clearance from these assays has been appreciated for almost 40 years (Gillette, 1963; Romer and Bickel, 1979; Obach, 1996;McLure et al., 2000). Consistent with established theory, correction for free fraction differences between microsomes and plasma has improved the predictive utility of microsomal data (Obach, 1996, 1997,1999; Obach et al., 1997; Carlile et al., 1999). In contrast, accurate predictions of hepatic clearance have also been made without the need for such a correction (Obach, 1999). In an analysis of 29 clinical compounds of known clearance by Obach et al., accounting for free fraction differences had the largest impact on acidic drugs (Obach, 1999). In contrast, the clearances of basic compounds were predicted with equivalent or improved accuracy by disregarding all protein binding data. In that data set, the free fraction differences between microsomes and plasma were 3 ± 1.6-fold for bases and 101 ± 148-fold for acids. When viewed in this manner, that data supports the general and intuitive concept that free fraction corrections are more likely to be required when differences in binding between microsomes and plasma are large. Since the extent of binding and the necessity of binding corrections are difficult to predict a priori, trends have been examined according to physiochemical properties (Obach, 1999). Regardless of these physiochemical properties, differences in the concentrations of nonspecific binding components among these matrices are expected to influence unbound fraction in a nonlinear manner (Romer and Bickel, 1979). When the overall extent of binding is low, less than proportional changes in free fraction are expected as the concentration of binding components are increased. As the average free fraction of the basic compounds in the study by Obach et al. was 32 ± 20%, the accuracy of predicted clearance in the absence of binding considerations may reflect this nonlinear relationship (Obach, 1999). In contrast, when binding is extensive, increases in the concentrations of nonspecific binding components within plasma alone may be expected to decrease free fraction in a linear manner (Romer and Bickel, 1979). Since bases can have microsomal free fractions much less than 10%, significant binding corrections may be required in these instances to accurately predict intrinsic clearance (McLure et al., 2000). When protein binding considerations are disregarded under these circumstances, even agreement of intrinsic clearance projections among individual microsomal assays may depend upon the coincidental presence of equivalent concentrations of nonspecific binding components. The potential impact of such a phenomenon is suggested by literature reports of microsomal protein concentrations varying as much as 200-fold among laboratories (Obach, 1999; Venkatakrishnan et al., 2000). As such, when nonspecific binding is extensive and binding considerations are disregarded, accurate predictions of in vivo clearance may be fortuitous. Therefore, the purpose of this work was to theoretically and experimentally illustrate the expected influence of microsomal concentration on estimates of fumic,1CLint,app, Km,app, and t1/2. The term microsomal concentration was used to represent the relative concentration differences in all binding components and enzymes used throughout the study. Concentration differences in both defined and undefined microsomal components were achieved through 5-fold differences in the total amount of microsomal protein added to the incubation mixture. Two compounds were chosen to represent situations where differences in the concentrations of nonspecific binding components are expected to have maximal (high nonspecific binding) and minimal (low nonspecific binding) influence on free fraction (Romer and Bickel, 1979).
Experimental Procedures
Materials.
Compounds A and B and all internal standards were obtained from Pfizer's proprietary sample bank (Groton, CT). The physiochemical properties of these two compounds are reported in Table1. All compounds were >99% pure, as determined by high-pressure liquid chromatography. Pooled human liver microsomes (n > 56) were obtained from an in-house bank of liver microsomes maintained at Pfizer Global Research and Development (Groton, CT). Microsomal CYP content, estimated as described previously (Omura and Sato, 1964), was 0.43 nmol/mg of protein. Solvents and other reagents were obtained from common sources and were of reagent grade or better. NADPH (97% purity) was obtained from Sigma Chemical Co. (St. Louis, MO).
Physiochemical properties of compounds A and B
Theoretical Impact of Microsomal Concentration on Free Fraction.
Assuming equilibrium kinetics and nonsaturable binding, the relationship between free fraction (fumic), dissociation constant (Kd), and binding component concentration (B) can be written in a manner consistent with that previously described in the literature (Romer and Bickel, 1979).
Theoretical Impact of Microsomal Concentration on Half-Life.
Assuming that only free substrate fumic · [S] is able to be acted upon by enzyme [E], and Vmax is expressed in terms of a catalysis rate constant (kcat) and the total mass of enzyme (Etotal), the velocity (v) is defined in eq. 3 by the Michaelis-Menten equation.
Theoretical Impact of Microsomal Concentration onKm.
The definition of apparent Km is provided in eq. 8.
Metabolic Incubations.
The microsomal clearances of compounds A and B were examined in pooled human liver microsomes at total protein concentrations of 0.46 and 2.3 mg/ml, corresponding to total CYP concentrations of 0.20 and 1.0 μM, respectively. These concentrations were achieved through dilution of a stock suspension of microsomes with sodium phosphate buffer (pH 7.4, 100 mM) to assure a 5-fold difference in the concentration of all nonspecific binding components. Selected substrate concentrations (0.1–300 μM) were incubated with magnesium chloride (5 mM) and NADPH (2 mM) in 0.75 ml of sodium phosphate buffer (pH 7.4, 100 mM) at 37°C. Microsomes were preincubated for approximately 5 min before the reaction was initiated by the addition of NADPH. Aliquots of 100 μl were removed from the incubations at 0, 5, 10, 15, 30, and 45 min and added to 100 μl of acetonitrile to terminate further metabolism. All incubations were conducted in triplicate.
Equilibrium Dialysis.
Compounds A and B (3.0 μM) were mixed with the indicated pooled human liver microsomes suspended in magnesium chloride (5 mM) and sodium phosphate buffer (pH 7.4, 100 mM). The mixture (1.0 ml) was subject to equilibrium dialysis versus 1.0 ml of sodium phosphate buffer (pH 7.4, 100 mM) at 37°C using a Spectrum semi-micro apparatus (Spectrum Industries, Los Angeles, CA) and Spectra-Por no.2 membranes with molecular weight cutoff of 12 to 14 kDa. Equilibrium was achieved by rotating the cells at 25 rpm for 4.5 h (Obach, 1997). All experiments were performed in triplicate.
High-Pressure Liquid Chromatography Analysis.
Quantitation of compound A was accomplished using an LC-MS assay. Samples were injected (1–5 μl; HTSPal Autosampler, CTC Analytics, Carrboro, NC) onto a Primesphere 2.0- × 30-mm 5-μm C18 column (Phenomenex, Torrance, CA) with a run time of 2.0 min. Analytes were eluted with acetonitrile/ammonium acetate (pH 6.8, 10 mM)/isopropanol (60:39:1, v/v/v) at a flow rate of 0.550 ml/min produced by a Shimadzu LC-10ADVP binary pump (Shimadzu, Kyoto, Japan). From 0.4 to 2.0 min the entire column effluent entered the Turbo Ionspray source (500°C, 7 liters/min nitrogen) of a PE-Sciex API-150 single quadrupole mass spectrometer (Concord, ON, Canada). Compound A and the internal standard were measured using single ion monitoring (m/z = 442.2 and 480.2, respectively) with positive ionization at a retention time of 0.7 min.
Quantitation of compound B was accomplished using an LC-MS/MS assay. Samples were injected (1–5 μl; HTSPal Autosampler) onto a monitor 3.0- × 30-mm 3-μM C18 column (Column Engineering, Ontario, Canada) maintained a 37°C with a run time of 4.5 min. Analytes were eluted with a high-pressure linear gradient program consisting of methanol (“A”) and ammonium acetate (pH 6.8, 10 mM)/isopropanol (99:1, v/v) (“B”) produced by two Shimadzu LC-10ADVP binary pumps and a 10-μl static mixer with a combined flow rate of 0.300 ml/min. An initial concentration of 20% A was immediately ramped to 100% A in 3 min and held for 0.5 min. The system was returned to the initial conditions in a single step and allowed to equilibrate for 1 min. From 1.5 to 4.0 min the entire column effluent entered the Turbo Ionspray source (500°C, 8 liters/min nitrogen) of a PE-Sciex API-3000 triple quadrupole mass spectrometer. Compound B and the internal standard were measured using multiple reaction monitoring (m/z = 411.2–339.2 and 466.1–380.1, respectively) with negative ionization at a retention time of 2.3 and 2.4 min, respectively.
Standard curves were used for analysis of equilibrium dialysis, and microsomal incubation samples were used for enzyme kinetic calculations. Equilibrium dialysis samples were prepared by mixing microsome samples with control buffer or buffer samples with control microsomes to yield an identical matrix, which were combined with internal standard in acetonitrile to precipitate proteins prior to analysis of supernatant. Samples from microsomal incubations were vortexed and centrifuged, and an aliquot of supernatant was combined with internal standard for analysis.
Calculations.
The initial reaction velocity (v0) was calculated by multiplying the nominal substrate concentration at time 0 ([S]0) by the slope of the log-linear regression from the concentration versus time relationship (k) and dividing by the protein concentration of the incubation, v0 = −k · [S]0/[P]. The velocity data were plotted in double reciprocal and Eadie-Hofstee plots to confirm enzyme kinetics consistent with a single metabolic site. Apparent Km andVmax were estimated by modeling velocity versus substrate concentration using the Michaelis-Menten equation and WinNonlin 2.1 (Pharsight Inc., Palo Alto, CA). In contrast, microsomal half-lives were calculated using the slope of the log-linear regression from the concentration versus time relationship (k),t1/2 = −ln(2)/k. Consistent with the required assumptions outlined in the theoretical section, half-life was reported for the lowest concentration examined (such that S ≪ Km). Intrinsic clearance was calculated from half-life using the following formula (Obach et al., 1997).
Results
Effect of Microsomal Concentration on Free Fraction (Table 2).
A 5-fold increase in microsomal concentration was associated with an equivalent 5-fold decrease in free fraction of compound A (high nonspecific binding) from 0.055 to 0.012 (Table2). In contrast, the free fraction of compound B (low nonspecific binding) was not proportionally influenced by microsomal concentration (Table 2).
Influence of 5-fold changes in protein and enzyme concentration on the estimated values of fumic, v, CLint, t1/2, and apparent Km
Effect of Microsomal Concentration on Half-Life and CLint,app (Table 2).
The half-life of compound A was independent of microsomal concentration. In this case, the decrease in CLint,app from 80 to 17 ml/min/kg was proportional to the increase in microsomal concentration. For compound B, the 5-fold increase in microsomal concentration was associated with a decrease in half-life from 19 to 5.9 min. In this case, there was no proportional change in CLint,app. Correction of CLint,app for the free fraction yielded estimates that were independent of microsomal concentration for both compounds.
Effect of Microsomal Concentration onKm,app.
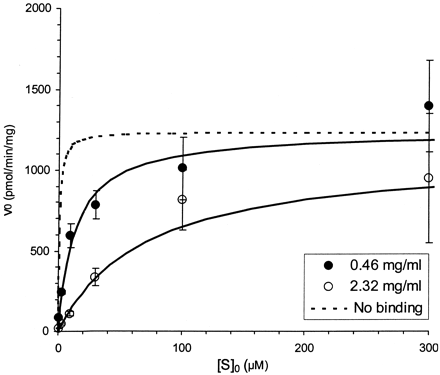
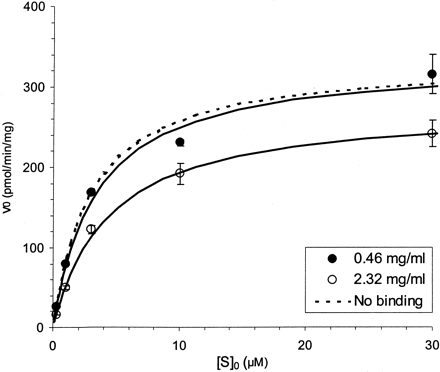
The Km,app of compound A increased proportionally to the 5-fold increase in microsomal concentration. In this case, a 5-fold increase in microsomal concentration was associated with an equivalent increase in Km,app from 13 to 70 μM (Fig. 1, Table 2). In contrast, the Km,app of compound B was not proportionally influenced by changes in microsomal concentration (Fig.2, Table 2). SinceVmax is normalized to total protein concentration it was unaffected for both compounds A and B (Fig. 2, Table 2). In addition, correction of Km,appfor free fraction yielded estimates that appeared to be independent of microsomal concentration for both compounds.
Saturation plot for compound A at two different microsomal protein (0.46 and 2.3 mg/ml) and enzyme (0.25 and 1.0 μM CYP) concentrations.
Solid lines represent the best fit to the data from individual runs using the Michaelis-Menten equation. The dashed line represents a simulation using the estimated parameters, following correction for nonspecific microsomal binding. Error bars represent standard deviation.
Saturation plot for compound B at two different microsomal protein (0.46 and 2.3 mg/ml) and enzyme (0.25 and 1.0 μM CYP) concentrations.
Solid lines represent the best fit to the data from individual runs using the Michaelis-Menten equation. The dashed line represents a simulation using the estimated parameters, following correction for nonspecific microsomal binding. Error bars represent standard deviation.
Discussion
Hepatic microsomal lability represents one of the simplest and most widely used tools to predict human clearance during the drug discovery process (Iwatsubo et al., 1997; Rodrigues, 1997; Obach, 1999). Two methods are commonly employed, including thet1/2 method, in which the first order rate of metabolism is determined at low substrate concentrations, and the enzyme kinetic method, in which Vmax andKm are determined (Rane et al., 1977; Obach et al., 1997). Estimates of in vivo intrinsic clearance are typically made through the use of scaling factors applied to the observed in vitro data on the basis of protein concentration (Houston, 1994). As previously reviewed, this use of microsomes requires many assumptions, including one that the substrate is freely available for metabolism (Houston and Carlile, 1997). Many compounds can bind nonspecifically to microsomes, thereby invalidating this assumption (Obach, 1996, 1997,1999; Obach et al., 1997; Venkatakrishnan et al., 2000). In these cases, increasing concentrations of nonspecific binding components within the incubation will decrease the apparent intrinsic clearance of compounds that bind to liver microsomes (Gillette, 1963; Obach, 1997;McLure et al., 2000). Despite the clear implications of these findings for scaling intrinsic clearance, methods disregarding all protein binding considerations can yield clearance projections of the same accuracy as those that account for the extent of plasma and microsomal binding (Obach, 1999). Discussion of this phenomenon in the literature frequently follows a delineation of the impact of binding corrections and experimental outcomes around physiochemical properties (Obach, 1999; McLure et al., 2000). Although the conclusions of these analyses may have empiric value, they do not directly relate the underlying factors known to influence free fraction to the experimental outcomes in a quantitative manner. As such, relationships between the necessity of nonspecific binding considerations and physiochemical properties should be tenuously considered when conducting studies to characterize additional compounds. Herein, we have theoretically and experimentally illustrated the expected impact of microsomal concentration on several parameters that are commonly reported as measures of microsomal lability. Projected and observed results were quantitatively compared in terms of -fold changes to fumic,t1/2, CLint,app, andKm,app. The theories were derived from very simple rearrangements of the relationship between free fraction and binding component concentration (Romer and Bickel, 1979). Two proprietary compounds were chosen to experimentally illustrate two extremes depicted by the derived theories, viz.; high nonspecific binding and low nonspecific binding. The highly bound representative (compound A, fumic = 0.055) was a basic and highly lipophilic compound, whereas the compound representative of low binding (compound B, fumic = 0.86) was a slightly less lipophilic acid (Table 1). Although the structures of these compounds are not revealed, we submit that the conclusions have general applicability to other compounds regardless of specific chemical structures or physiochemical characteristics.
High Nonspecific Binding.
A 5-fold increase in microsomal concentration was associated with a proportional decrease in the free fraction of compound A (Table 2). No difference in metabolic half-life was observed; however, scaling of the observed microsomal t1/2 to an estimate of apparent intrinsic clearance resulted in a 5-fold lower value at the higher microsomal concentration (Table 2). Consistent with this result, the Km,app of compound A increased proportionally to the increase in microsomal concentration (Table 2, Fig. 1). Each of these observations is consistent with the expected behavior for a highly bound compound predicted by eqs. 2, 7, and 9. We conclude that the 5-fold difference in CYP concentration is offset by an inversely proportional change in free fraction. This conclusion was supported when correction for the free fraction available for metabolism resulted in virtually identical estimates of intrinsic clearance under both conditions (Table 2). Although these findings are in general agreement with recent simulations published by McLure et al. (2000), the lack of a direct quantitative relationship between these variables in that study prevents a detailed comparison. We expect that this class of compounds will frequently require the correction factor previously proposed by Obach (1996) for free fraction differences between plasma and microsomes to accurately predict clearance in vivo. In this circumstance, the quantitative differences in protein and lipid content between microsomes and plasma alone could reasonably be expected to result in significant differences in the free fraction between these two matrices. Since the free fraction in microsomes is determined, in part, by the choice of microsomal concentration in the incubation, equivalent free fractions between plasma and microsomes should be considered coincidental. It is therefore unlikely that the clearance of these compounds will be accurately predicted by disregarding all binding. These data also suggest that one could avoid indiscriminate and labor intensive protein binding studies by determining the metabolic half-life at two different microsomal concentrations. If half-life is unchanged, then the compound may be highly bound. If half-life is changed proportionally then the compound is expected to have a low extent of nonspecific binding as described below.
Low Nonspecific Binding.
Less than proportional changes in the free fraction of compound B were observed relative to microsomal concentration (Table 2). It is important to note that a nonlinear decrease in free fraction was observed at the higher microsomal concentration. This is consistent with eq. 1, which predicts that compounds with high free fractions will be influenced nonlinearly by changes in the concentrations of nonspecific binding components. At 5-fold higher microsomal concentration, the t1/2 of compound B was decreased by greater than 3-fold (Table 2). The slight difference in free fraction, observed in this case, contributes to the slightly less than proportional difference in t1/2 (Table2). Scaling of the observed microsomal t1/2to CLint,app, resulted in estimates that were relatively independent of microsomal concentration (Table 2). Consistent with this result, the Km,app of compound B was also relatively independent of microsomal concentration (Table 2, Fig. 2). Again, the slight difference in free fraction may account for the small differences in CLint,appand Km,app. This is evidenced by the even smaller differences in these parameters when corrected for free fraction (Table 2). Each of these observations is consistent with the expected behavior predicted by eqs. 2, 7, and 9 for a compound with low nonspecific binding. In this case, we conclude that the increase in CYP afforded by increasing the microsomal concentration within the incubation is accomplished without a significant effect on the free fraction of substrate available for metabolism. Becauset1/2 from the microsomes decreases proportionally, scaling of intrinsic clearance results in estimates that are independent of microsome concentration. We expect that this class of compounds may not require the correction factor previously proposed by Obach (1996) for free fraction differences between plasma and microsomes to accurately predict clearance in vivo. However, it is important to note that highly free compounds in microsomes may still be highly bound to plasma proteins. In such cases, it may be possible to ignore in vitro microsomal protein binding but necessary to account for plasma protein binding when predicting clearance.
In summary, we have derived a series of companion equations that quantitatively illustrate the combined influences of nonspecific binding component and enzyme concentration on several relevant kinetic endpoints in a manner that is intuitively understood. Two compounds were chosen to experimentally illustrate the expected results under two distinct situations, namely a high and low degree of nonspecific microsomal binding. These results illustrate that, regardless of physiochemical properties themselves, compounds with a high degree of nonspecific binding to microsomes are likely to require additional considerations for free fraction when predicting clearance in vivo. We believe this work offers additional theoretical and practical clarity to the appropriate design and interpretation of microsomal lability assays.
Footnotes
- Abbreviations used are::
- fumic
- microsomal fraction unbound
- CLint,app
- apparent intrinsic clearance
- CYP
- cytochrome P450
- Km,app
- apparent Michaelis-Menten constant
- Received April 4, 2001.
- Accepted July 19, 2001.
- The American Society for Pharmacology and Experimental Therapeutics