Abstract
A method for the simultaneous evaluation of the activities of seven major human drug-metabolizing cytochrome P450s (CYP3A4, CYP2D6, CYP2C9, CYP1A2, CYP2C19, CYP2A6, and CYP2C8) was developed. This method uses an in vitro cocktail of specific substrates (midazolam, bufuralol, diclofenac, ethoxyresorufin, S-mephenytoin, coumarin, and paclitaxel) and fast gradient liquid chromatography tandem mass spectrometry. The assay incubation time is 20 min, which is in the linear range for all of the substrates, and the analysis time is 4 min/sample. Substrate specificity was confirmed by incubatingEscherichia coli-expressed enzymes with the cocktail. Potent specific inhibitors of the seven enzymes (ketoconazole, quinidine, sulfaphenazole, tranylcypromine, quercetin, furafylline, and 8-methoxypsoralen) were evaluated in cocktail and individual substrate incubations. Five of these inhibitors were further studied to determine more precise IC50 values for inhibition of the seven enzymes. The IC50 values obtained in both cocktail and individual incubations were in good agreement with published values. This cocktail method offers an efficient, robust way to determine the cytochrome P450 inhibition profile of large numbers of compounds. The enhanced throughput of this method greatly facilitates its use to assess CYP inhibition as a drug candidate selection criterion.
Compounds that are potent inhibitors of one or more cytochrome P450 enzymes have a potential for drug-drug interactions. Potent drug-drug interactions can result in serious side effects (e.g., ketoconazole and terfenadine) (Honig et al., 1993). As a result, preclinical (in vitro) and some clinical (in vivo) interaction studies are now important components of the drug candidate selection process (Honig et al., 1993; Peck et al., 1993; Ball et al., 1997; Ko et al., 1997). In vitro studies to determine IC50 or Kivalues for inhibition of the major human drug-metabolizing enzymes are currently very time consuming using traditional methods (e.g., HPLC3 and fluorescent enzyme assays), which permit the evaluation of the activity of a single enzyme at one time (Wester et al., 2000). The use of high-throughput compound screening methods and combinatorial chemistry in the discovery of new chemical entities has led to a great increase in the number of compounds to be evaluated for CYP inhibition. The traditional enzyme assays are cumbersome for quickly evaluating large numbers of compounds.
Recently, several articles have been published outlining higher throughput, in vitro cytochrome P450 assays for the evaluation of putative inhibitors. These assays use a variety of techniques and substrates (e.g., mass spectrometry, radioactive/fluorescent probes). The fluorescent and radioactive assays require the use of expressed CYP enzymes instead of human liver microsomes for at least some of the isozyme assays due to the poor specificity of the substrates used. Interference from the putative inhibitor, the necessity of time-consuming sample preparation steps (e.g., solid phase extraction), and the inability to assay multiple enzymes in a single sample are other limitations of these methods (Crespi et al., 1997; Ayrton et al., 1998; Moody et al., 1999).
This article outlines a new, robust method for the simultaneous evaluation of the activities of seven major human drug-metabolizing enzymes (CYP3A4, CYP2D6, CYP2C9, CYP1A2, CYP2C19, CYP2A6, and CYP2C8). This method uses an in vitro cocktail of specific, well characterized CYP probe substrates to monitor enzyme activity and inhibition. Sample analysis is by fast gradient LC-MS/MS with SRM of the specific metabolites. Data on well characterized CYP inhibitors demonstrate that this method can be used to rapidly predict potent CYP inhibition (IC50 values less than 10 μM) and to quickly determine more precise IC50 values that are in agreement with literature values.
Experimental Procedures
Chemicals.
S-Mephenytoin, furafylline, and ketoconazole were obtained from Salford Ultrafine Chemicals (Manchester, UK). Tris base, dibasic potassium phosphate, and magnesium chloride hexahydrate were purchased from J.T. Baker (Phillipsburg, NJ). Tranylcypromine and 8-methoxypsoralen were obtained from Aldrich Chemical Company (Milwaukee, WI). Ethoxyresorufin was from Molecular Probes, Inc. (Eugene, OR). Bufuralol and dextrorphan were purchased from Gentest (Woburn, MA). Quercetin dihydrate and formic acid were obtained from Fluka Biochemika (Milwaukee, WI). Midazolam was obtained from the Wyeth-Ayerst Compound Room (Princeton, NJ). Concentrated HCl was from VWR (Philadelphia, PA). All other chemicals were purchased from Sigma Chemical Company (St. Louis, MO).
Microsome Preparation.
Human liver microsomes were prepared by differential ultracentrifugation from donor human liver samples obtained from IIAM (Scranton, PA) according to published protocols (Lake, 1987). Microsomal cytochrome P450 content was determined by CO-difference spectrum (Omura and Sato, 1964). Protein content was determined by the Bradford method (Bradford, 1976; Macart and Gerbaut, 1982). Microsomes used in these studies were pooled from a minimum of five individuals.
Cytochrome P450 Expression in Escherichia coli and Membrane Preparation.
E. coli expression plasmids (modified pCW, a common CYP expression plasmid) containing human NADPH-cytochrome P450 reductase and human CYP3A4, CYP2D6, CYP2C9, CYP1A2, CYP2C19, CYP2A6, and CYP2C8 were provided by the LINK industrial consortium, an academic and industrial collaboration. The constructs are described and the enzymes were expressed and prepared essentially as outlined in published protocols (Gillam et al., 1993; Blake et al., 1996; Pritchard et al., 1997, 1998; Li et al., 1999). Protein and cytochrome P450 concentrations were determined using published protocols (Omura and Sato, 1964; Bradford, 1976; Macart and Gerbaut, 1982).
Assay Incubation Conditions.
All incubations (individual substrate and cocktail) were done under conditions shown to be linear with respect to time, protein concentration, and substrate concentration (all at the apparentKm concentration). Incubations contained either the cocktail of probe substrates or an individual substrate (Table 1). Each sample contained 0.5 mg/ml human liver microsomes, 10 mM MgCl2, 100 mM potassium phosphate buffer (pH 7.4), and probe cocktail or individual substrate in a total volume of 0.5 ml. Samples were preincubated for 5 min at 37°C in a shaking water bath. The reactions were initiated by addition of NADPH to a final concentration of 2 mM. Incubations were carried out for 20 min and terminated by addition of 0.25 ml of acetonitrile and 0.5 μM dextrorphan as an internal standard. Samples were centrifuged at 3000 rpm for 10 min at 4°C to pellet the precipitated protein. The acetonitrile supernatant was evaporated under a stream of nitrogen (approximately 30 min). The aqueous samples were transferred to HPLC vials for analysis.
Probe substrates, metabolites, SRM (precursor and product), and collision energies of the probe substrate cocktail
LC-MS/MS Method.
Samples were analyzed by LC-MS/MS in APCI positive mode using a Hewlett Packard (Palo Alto, CA) 1090 HPLC coupled to a Finnigan (San Jose, CA) TSQ 7000 mass spectrometer using a modification of a published protocol (Ayrton et al., 1998).
A Keystone (Bellefonte, PA) BDS Hypersil C8 column (20 × 2 mm, 5 μm) was used for the separation. A two-component mobile phase, pumped at 1 ml/min, contained the following: solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid in acetonitrile). A gradient of solvent B from 2 to 95% over 4 min was applied on the column and then cycled back to the initial condition. Column temperature was maintained at 50°C, and the sample temperature was kept at 5 to 8°C.
The positive ion APCI mass spectra were acquired by flow injection of 80 μl of the samples. The sample was desolvated at a capillary temperature of 170°C and vaporizer temperature of 500°C. The sheath gas was set at 90 psi, and the flowmeter reading of auxillary gas was set at 30. The APCI needle voltage, the electron multiplier, and the conversion dynode were set at 4.5, 1700, and 15 kV, respectively. The mass scan used SRM for each metabolite at a scan rate of 0.1 s/scan (Table 1; Fig. 1). The data acquisition was performed on a Finnigan TSQ 7000 APII mass spectrometer equipped with a Digital DEC station computer and software of ICL 8.3.2 and ICIS 8.3.0 (Finnigan). Data were analyzed by LCQuan version 1.2 (Finnigan).
Elution profiles of the metabolites using the LC-MS/MS method.
A, 7-hydroxycoumarin, retention time 1.60 min; B, resorufin, retention time 1.97 min; C, 4′-hydroxymephenytoin, retention time 1.78 min; D, dextrorphan, retention time 1.89 min; E, 1′-hydroxybufuralol, retention time 1.90 min; F, 4′-hydroxydiclofenac, retention time 2.52 min; G, 1′-hydroxymidazolam, retention time 2.24 min; H, 6α-hydroxypaclitaxel, retention time 2.75 min.
Substrate Specificity Determination.
The specificity of each substrate for its selected enzyme was evaluated by performing incubations with the probe cocktail as described above but substituting 50 pmol/ml E. coli-expressed CYP3A4, CYP2D6, CYP2C9, CYP1A2, CYP2C19, CYP2A6, or CYP2C8 for the human liver microsomes. Samples were analyzed for specific product formation by LC-MS/MS as described above.
Comparison of the Utility of the Cocktail and the Individual Substrates for Screening for Potent CYP Inhibition (IC50 < 10 μM).
Well characterized inhibitors of specific CYP enzymes (ketoconazole/CYP3A4, quinidine/CYP2D6, sulfaphenazole/CYP2C9, tranylcypromine/CYP2C19, quercetin/CYP2C8, furafylline/CYP1A2, and 8-methoxypsoralen/CYP2A6) were incubated at a concentration of 10 μM with both the cocktail and individual substrates alone to determine whether strong inhibition could be detected and whether the cocktail and individual incubations would yield similar results. Incubations were performed (as described above) with 10 μM inhibitor, human liver microsomes, and either the cocktail or individual substrates alone. Furafylline and 8-methoxypsoralen were preincubated for 5 min at 37°C with 2 mM NADPH and human liver microsomes before addition of the cocktail or individual substrate to initiate the reaction. For all of the inhibitors, a comparison was made to the activity of control incubations that did not contain the inhibitor. Conclusions were made as to whether the IC50 values were greater than or less than 10 μM.
IC50 Determination.
To further validate the utility of the probe substrate cocktail in the assessment of CYP inhibition, IC50 values were determined for the inhibition of the CYP enzymes by ketoconazole, quinidine, sulfaphenazole, tranylcypromine, and quercetin. Human liver microsomal incubations were done with the cocktail and the individual substrates as described above with the exception that they also included 0, 0.1, 1, 10, or 50 μM inhibitor. Comparison was made to control incubations (0 μM inhibitor) and activity expressed as the percentage of control activity remaining. IC50values were determined by linear interpolation.
Results
Substrate Specificity.
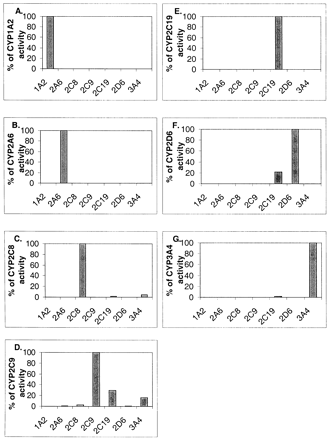
Figure 2 illustrates the results of incubating the probe substrate cocktail with E. coli-expressed CYP3A4, CYP2D6, CYP2C9, CYP1A2, CYP2C19, CYP2A6, and CYP2C8. In each case, the substrate is metabolized exclusively or primarily by its specific enzyme. Multiple enzymes metabolize paclitaxel, diclofenac, bufuralol, and midazolam, but the activity levels are less than 30% of the activity of the enzyme for which the substrate is a probe. In most cases, the activity levels of the multiple enzymes are less than 10% of the activity of the main enzyme. These results indicate that the substrates are specific for the desired enzyme.
Specificity of the probes in the cocktail for catalysis by CYP enzymes.
Incubations with the cocktail and E. coli-expressed CYP3A4, CYP2D6, CYP2C9, CYP1A2, CYP2C19, CYP2A6, and CYP2C8 were performed as described under Experimental Procedures. Activity is expressed as a percentage of the activity obtained for the desired isozyme. The substrates are ethoxyresorufin (A), coumarin (B), paclitaxel (C), diclofenac (D), S-mephenytoin (E), bufuralol (F), and midazolam (G).
Comparison of the Utility of the Cocktail and the Individual Substrates for Screening for Potent CYP Inhibition (IC50 < 10 μM).
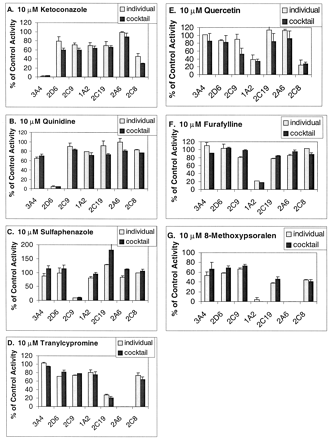
Figure 3 shows the results of incubations of the individual substrates and the cocktail with human liver microsomes in the presence of 10 μM ketoconazole, quinidine, sulfaphenazole, tranylcypromine, quercetin, furafylline, and 8-methoxypsoralen. In each case, there is close agreement between the individual incubations and the cocktail incubations. The current data (from both cocktail and individual incubations) demonstrate that ketoconazole is a potent inhibitor of CYP3A4 and CYP2C8, quinidine is a potent inhibitor of only CYP2D6, sulfaphenazole of only CYP2C9, tranylcypromine of CYP2C19 and CYP2A6, quercetin of CYP1A2 and CYP2C8, furafylline of CYP1A2, and 8-methoxypsoralen of CYP2C19, CYP2C8, CYP2A6, and CYP1A2.
Comparison of the cocktail and the individual substrates for screening putative inhibitors for potent CYP inhibition (IC50 < 10 μM).
Incubations were done with 10 μM specific inhibitors (ketoconazole, quinidine, sulfaphenazole, furafylline, tranylcypromine, 8-methoxypsoralen, and quercetin), the cocktail or individual probes, and human liver microsomes as described under Experimental Procedures. Activity is expressed as the percentage of activity remaining compared with a control sample containing no inhibitor. Activities are means of duplicate incubations.
IC50 Determinations.
The IC50 values determined using both the cocktail and the individual probe substrates in incubations containing four or five concentrations of ketoconazole, quinidine, sulfaphenazole, tranylcypromine, and quercetin are listed in Table2.
Comparison of IC50 values obtained using the cocktail and individual probe substrates
For ketoconazole, both the cocktail and individual incubations predict strong inhibition of CYP3A4 and CYP2C8. No inhibition of CYP2A6 was observed in either incubation, whereas moderate inhibition of CYP2D6, CYP2C9, CYP1A2, and CYP2C19 was observed in both sets of incubations.
For quinidine, both the individual and cocktail incubations predict an IC50 value of less than 0.1 μM for inhibition of CYP2D6. Moderate inhibition of CYP3A4 was observed in both sets of incubations. There was only weak or no significant inhibition for the rest of the enzymes using either method.
For sulfaphenazole, both the cocktail and individual incubations predict an IC50 value of 1 μM for inhibition of CYP2C9. No other enzymes were significantly inhibited in either individual or cocktail incubations.
For tranylcypromine, both the cocktail and individual incubations predict strong inhibition of CYP2C19 and CYP2A6. The remaining enzymes were either moderately or weakly inhibited in both sets of incubations.
For quercetin, both the cocktail and individual incubations predict strong inhibition of CYP2C8 and CYP1A2. No significant inhibition was observed in either individual or cocktail incubations for CYP3A4, CYP2D6, CYP2C19, or CYP2A6. Moderate to weak inhibition of CYP2C9 was observed in both the cocktail and individual incubations.
Discussion
The objective of this study was to develop an increased throughput method for evaluating the inhibition of the major human drug-metabolizing cytochrome P450s. This was achieved by developing an in vitro cocktail of seven probe substrates that can be incubated with human liver microsomes or expressed CYPs and analyzed by fast gradient LC-MS/MS (Table 1; Fig. 1). The run time for analysis of a sample is 4 min.
The present assay using a cocktail of probe substrates offers many advantages over previously published methods for evaluation of cytochrome P450 activity and inhibition (Crespi et al., 1997; Ayrton et al., 1998; Moody et al., 1999). The substrates, which are well characterized CYP substrates, are very specific for their respective enzymes (Fig. 2). Similar specificity results have been previously reported (Mancy et al., 1999; Mankowski, 1999; Masimirembwa et al., 1999). Because of the high substrate specificity, human liver microsomes, as well as expressed CYPs, can be used in all assays. All of the substrates and metabolites are readily available commercially; no radioactivity is required, and interference with or quenching of assays has not been observed. SRM of the specific metabolites by LC-MS/MS ensures that only one product is monitored for each substrate. Sample preparation before analysis is minimal and compatible with large numbers of samples due to the specificity of the analytical method. Our studies have also shown that samples are stable at 4°C for at least 4 days after preparation (data not shown), which also is an advantage when generating large numbers of samples. One of the biggest advantages is the greatly increased throughput achieved by performing a single incubation and analysis for seven different enzymes. By conducting seven assays in a single sample, the amount of human liver microsomes, potential inhibitor, and NADPH required for assays is decreased. Further experiments indicate that sample volume can be decreased to between 100 and 200 μl, which would further decrease the amount of microsomes and inhibitor necessary for experiments. Additionally, this method is also readily compatible with automation. Experiments have been done successfully in 96-well plates using liquid handlers, an automated workstation (for incubations), and a 96-well autosampler (data not shown). As many as 47 compounds could be evaluated for potent CYP inhibition (10 μM incubations) or IC50values determined for 12 compounds in a single plate.
A compound that inhibits an enzyme with an IC50of less than 10 μM is generally considered to be a strong inhibitor of that isozyme. Historically, compounds with IC50 values in this range (and lower) tend to have more drug-drug interaction concerns (Honig et al., 1993; Peck et al., 1993; Ball et al., 1997; Ko et al., 1997). The experiments incubating the cocktail with 10 μM of the specific inhibitors (e.g., ketoconazole) and those incubating the cocktail with four or five concentrations of the specific inhibitors demonstrate that the present method can be used both to screen for potent inhibition and to rapidly evaluate the IC50 values. There is good agreement between the values obtained in cocktail and individual substrate incubations, although the absolute enzyme rates vary between the cocktail and individual substrate incubations for some enzymes. Typical control specific activities obtained using the cocktail are 121 ± 7 pmol/min/mg of protein for CYP3A4, 45.9 ± 0.3 pmol/min/mg for CYP2D6, 241 ± 3 pmol/min/mg for CYP2C9, 53.2 ± 0.8 pmol/min/mg for CYP1A2, 2.9 ± 0.3 pmol/min/mg for CYP2C19, 89.1 ± 0.9 pmol/min/mg for CYP2A6, and 27 ± 1 pmol/min/mg for CYP2C8. Specific activities in individual substrate incubations are the same as in the cocktail for CYP1A2 and CYP2A6, 15 to 20% greater than the cocktail values for CYP2D6, CYP2C9, CYP2C19, and CYP2C8, and 30 to 40% greater for CYP3A4. The IC50predictions from the rapid screen and the IC50evaluations agree with each other and with published literature values for most of the enzymes and inhibitors explored (Sousa and Marletta, 1985; Pasanen et al., 1988a,b; Gascon and Dayer, 1991; Kerry et al., 1994; Rahman et al., 1994; Baldwin et al., 1995; Ching et al., 1995;Newton et al., 1995; Bourrie et al., 1996; Jones et al., 1996; Mancy et al., 1996; Ono et al., 1996; Crespi et al., 1997; Crespi and Penman, 1997; Eagling et al., 1998; Hickman et al., 1998; Jung et al., 1998;Moody et al., 1999). The one exception to this agreement with published values is the inhibition of CYP2C9 by tranylcypromine. Little has been published specifically about this relationship; however, Crespi et al. (1997) have reported the inhibition of CYP2C9 by tranylcypromine is comparable with that of CYP2C19. We see significantly greater inhibition of CYP2C19 over CYP2C9 in both cocktail and individual incubations (Table 2). This could be due to the use of different substrates or expressed enzymes versus human liver microsomes in the two studies. Nonetheless, these results demonstrate that four or five inhibitor concentrations are generally adequate for evaluating IC50 values. It is possible also that the cocktail (or a subset thereof) could be used to quickly determineKi values for inhibition of multiple enzymes and to evaluate metabolism-dependent inhibition.
In conclusion, this article has demonstrated that the present cocktail can be used effectively to rapidly assess the cytochrome P450 inhibitory profile of new chemical entities. The method is robust and readily adaptable to automation (liquid handlers, 96-well plates, 96-well plate autosamplers) and small volumes (100–200 μl). It uses well characterized, readily available CYP substrates that are very specific for the particular enzyme probed. Either human liver microsomes or expressed cytochrome P450s can be used for all assays, and the simultaneous assay of seven enzymes in a single small-volume sample conserves both microsomes/enzymes and putative inhibitors (both of which may be limited in quantity). No radioactivity is used, and no interference by potential inhibitors was observed. This cocktail also has the potential to be used in evaluation of CYP induction/activation, rapid characterization of microsomal banks, rapid phenotyping of tissue (hepatic and extrahepatic) samples, and evaluation of hepatocyte/tissue slice enzyme activity. In addition, a similar cocktail could be developed to assess activity/inhibition of the phase II drug-metabolizing enzymes, although specific substrates for many of these enzymes are not yet known.
Footnotes
-
Send reprint requests to: Elizabeth Dierks, Wyeth-Ayerst Research, CN 8000, Princeton, NJ 08543-8000. E-mail:dierkse{at}war.wyeth.com
-
↵1 Portions of this article were previously presented at the 13th International Symposium on Microsomes and Drug Oxidations, Stresa, Italy, July 2000.
-
↵2 Current address: Metabolism and Pharmacokinetics, Bristol-Myers Squibb Pharmaceutical Research Institute, Wallingford, CT 06492-7600.
- Abbreviations used are::
- HPLC
- high-performance liquid chromatography
- CYP
- cytochrome P450
- LC-MS/MS
- liquid chromatography tandem mass spectrometry
- SRM
- selective reaction monitoring
- APCI
- atmospheric pressure chemical ionization
- Received August 14, 2000.
- Accepted October 9, 2000.
- The American Society for Pharmacology and Experimental Therapeutics