Abstract
An exhaustive real-time reverse transcriptase-polymerase chain reaction (PCR) quantification method was used to determine 15 of the catalytically active human UDP-glucuronosyltransferase (UGT) isoforms (1A1, 1A3, 1A4, 1A5, 1A6, 1A7, 1A8, 1A9, 1A10, 2B4, 2B7, 2B10, 2B11, 2B15, and 2B17). The specific primers for respective human UGTs were developed for differential determination. The cDNA derived from the 1A7 isoform was detected in the esophagus, the 1A8 and 1A10 isoforms were detected in the small intestine, and all other isoforms were detected in at least the liver by PCR. In all cases, single bands of the expected size on the agarose gel were confirmed to correspond with the predicted UGT isoform sequences. Each calibration curve showed linearity between the PCR crossing point and the calibrator copy number. The correlation coefficients were greater than 0.9957 with high reproducibility. This exhaustive measurement method was applied to UGT expression in 23 human tissue types. UGT was mostly expressed in the alimentary system and liver. We were surprised to find that extremely high expression in the liver was found for UGT2B4 and UGT2B15, which had, respectively, 8.98 and 4.38 times greater expression than UGT2B7 in the liver. In addition, even though expressed at low levels, several UGT isoforms were expressed in steroidogenic tissues, such as the breast, prostate, heart, and adrenal. Therefore, this quantification method may provide valuable information about the medical efficacy or pharmacokinetic characteristics of a wide variety of UGT-metabolized drugs.
UDP-glucuronosyltransferase (UGT) is a member of the superfamily of endoplasmic reticulum-bounded enzymes that catalyze glucuronidation of endogenous substrates, such as bilirubin, bile acids, and steroids in addition to a wide range of exogenous xenobiotics (Wells et al., 2004; Sun et al., 2006). In humans, 31 genes, including some pseudogenes, have been identified to date (Tukey and Strassburg, 2000; Mackenzie et al., 2005). Of these, 17 isoforms belonging to either the UGT1 (1A1, 1A3, 1A4, 1A5, 1A6, 1A7, 1A8, 1A9, and 1A10) or UGT2 (2A1, 2B4, 2B7, 2B10, 2B11, 2B15, 2B17, and 2B28) family seem to be catalytically active isoforms.
UGTs are involved in the metabolism of a wide variety of aglycones for both endogenous compounds and xenobiotics, and quantitative determination of these molecules will provide useful information about pharmacokinetics and adverse or beneficial effects of related drugs. For example, reducing the transcriptional activity of UGT1A1, which leads to the reduction of enzymatic activity of UGT1A1, exerts a strong positive influence on the toxicity of irinotecan, an antineoplastic agent, in the patient (Ando et al., 2005). However, no exhaustive investigations on the expression of these 17 catalytically active isoforms in various human tissues have been reported until now, probably due to difficulties with carrying out differential determination because of the sequence similarities. The UGT1A subfamily genes have multiple exon 1 structures, and exons 2 to 5 are shared by all UGT1A isoforms and are joined to exon 1 in the mature transcripts (Zhang et al., 2004). Multiple sequence alignment for exon 1 of UGT1A3, 1A4, and 1A5 showed between 86 and 92% identity and that of UGT1A8, 1A9, and 1A10 showed 75 and 91% identity (Gong et al., 2001). On the other hand, the UGT2B subfamily isoforms are encoded individually and consist of six exons (Mackenzie et al., 2005), with higher identity (84–97%; ClustalW, http://www.ebi.ac.uk/Tools/clustalw2/index.html) than that for members of the UGT1A subfamily. The amino acid UGT1A and 2B subfamily sequences have also been reported to have a high percent identity (Tukey and Strassburg, 2000).
Numerous studies on the expression and localization of UGTs have been reported; most used reverse transcription (RT)-polymerase chain reaction (PCR) or Northern blotting (Beaulieu et al., 1996, 1998; Lévesque et al., 1997, 1999; Strassburg et al., 1997a,b, 1998, 2000; Tchernof et al., 1999; Tukey and Strassburg, 2001; Turgeon et al., 2001; Girard et al., 2003; Nakamura et al., 2008) and a few used Western blotting (Pillot et al., 1993; Ikushiro et al., 2006). In general, RT-PCR is a simple, yet sensitive, tool for detecting transcription products, whereas Western blotting can be used to verify the expression of translated proteins but requires specific antibodies for each target protein. However, both of these methods are unsuccessful in estimating the difference in expression levels between different target molecules because the efficiency of detection specificity varies with each target.
In a recent study, UGT1A4, 2B7, and 2B10, which are involved in nicotine glucuronidation, were analyzed by real-time PCR in eight human tissues, including the liver, kidney, small intestine, colon, stomach, trachea, lung, and brain (Kaivosaari et al., 2007). Because reverse transcription real-time PCR can be used to quantify the mRNA expression level relative to the copy number, this method can provide a precise, direct comparison between numerous different targets. Therefore, a real-time RT-PCR quantification method targeting 15 of the 17 catalytically active human UGT transcripts (excluding UGT2A1 and UGT2B28) was applied to precisely determine the relative mRNA expression levels of UGTs in various human tissues.
Materials and Methods
Preparation of Calibrators. Human total RNA of the liver (from a 69-year-old man who died of an intracranial hemorrhage) and small intestine (from a 46-year-old white male who died of a cerebrovascular accident) was purchased from Ambion (Austin, TX) and esophagus was obtained as part of the FirstChoice Human Total RNA Survey Panel (Applied Biosystems/Ambion Japan, Tokyo, Japan); the donor information is shown in Table 1. Reverse transcription was carried out using Illustra Ready-To-Go RT-PCR beads (GE Healthcare, Little Chalfont, Buckinghamshire, UK) according to the manufacturer's recommendations using 1 μg of total RNA as the template and 0.5 μg of oligo(dT)12–16 as the RT primer. The transcripts encoding members of the UGT1A subfamily (1A1, 1A3, 1A4, 1A5, 1A6, 1A7, 1A8, 1A9, and 1A10) were amplified by RT-PCR using the Gene Taq DNA polymerase (Nippon-Gene, Tokyo, Japan) with specifically designed forward primers as listed in Table 2 and the consensus reverse primer (5′-ATTCCATGTTCTCCAGAAGCA-3′), which was prepared from the consensus of exon 2 sequences from the reverse transcripts of three different tissues (esophagus cDNA for UGT1A7, small intestine cDNA for UGT1A8 and 1A10, and liver cDNA for all other members of the UGT1A subfamily). The transcripts for each member of the UGT2B subfamily (2B4, 2B7, 2B10, 2B11, 2B15, and 2B17) were amplified by RT-PCR using specific forward and reverse primers from the reverse transcribed human liver cDNA library. (The primer sequences of all UGT2B subfamily cDNAs for preparing calibrators, which were the same as the quantification primers, are listed in Table 2.) The housekeeping gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), was used as and internal control and was amplified with a specific primer pair (5′-ATGGGGAAGGTGAAGGTCG-3′/5′-TTACTCCTTGGAGGCCATGT-3′) to produce a 1008-bp amplicon from the human liver cDNA library. All amplicons were cloned into the pGEM-T Easy vector (Promega, Madison, WI) according to the manufacturer's recommendations. For the calibrators, each vector was transformed into Escherichia coli competent cells (JM109) and purified by the PureYield Plasmid Midiprep system (Promega), and then products were confirmed with sequencing by the dye terminator method, using an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems Japan, Tokyo, Japan).
Donor information
Human total RNAs from breast, adrenal, cerebellum, and stomach were derived from a single donor. Others samples were of pooled human total RNAs derived from three donors who are described in the table.
Primer sequence used for real-time RT-PCR quantification and for calibrator preparation
Real-Time PCR Conditions. Quantification of human UGT mRNA (National Center for Biotechnology Information Accession Number is shown in parentheses) for UGT1A1 (NM_000463), 1A3 (NM_019093), 1A4 (NM_007120), 1A5 (NM_019078), 1A6 (AY435141), 1A7 (NM_019077), 1A8 (NM_019076), 1A9 (NM_021027), 1A10 (NM_019075), 2B4 (NM_021139), 2B7 (NM_001074), 2B10 (NM_001075), 2B11 (NM_001073), 2B15 (NM_001076), 2B17 (NM_001077), and GAPDH (NM_002046) was performed using the LightCycler 480 system with a 96-well format (Roche Diagnostics, Tokyo, Japan) in a total reaction volume of 20 μl/well of LightCycler 480 SYBR Green I Master solution (Roche Diagnostics). The reaction mixture consisted of 0.5 μM concentrations of each of the specific primer pairs for quantification and 5 μl of a 6-fold diluted human cDNA library, which was reverse transcribed from human total RNA of various tissues. A serial diluted calibrator vector (1 ng–512 ag for UGTs or 0.5 ng–256 ag for GAPDH) for a standard curve and sterilized water for a negative control in 1× LightCycler 480 SYBR Green I Master solution were prepared. PCR amplification consisted of an initial 5-min denaturation step at 95°C, followed by 45 cycles of denaturation at 95°C for 10 s, annealing at 60°C for 12 s, and extension at 72°C for 1 min/1-kilobase pair amplification. Specificity of the amplified PCR product was assessed by performing a melting curve analysis on the LightCycler 480 system. The resulting melting curves allowed for discrimination between primer-dimers and specific products. The primer pair sequences that were used for real-time PCR detection for 15 kinds of human UGTs and GAPDH and the amplicon sizes are summarized in Table 2. The results of quantification were calculated as copy number ratio to GAPDH mRNA using the average molecular weight of one base-paired nucleotide (660 g/mol) and Avogadro's constant (6.022 × 1023).
Human Total RNA Samples and Reverse Transcription. Human total RNA of 19 samples was obtained from the FirstChoice Human Total RNA Survey Panel, which consists of pooled total RNA from 20 different normal human tissues (liver, esophagus, small intestine, colon, brain, thyroid, thymus, testes, prostate, ovary, placenta, cervix, heart, lung, trachea, kidney, bladder, spleen, skeletal muscle, and adipose tissue) from at least three tissue donors (Applied Biosystems Japan/Ambion). Total RNA from four other human tissues [cerebellum, adrenal gland, and breast (Ambion) and stomach (Clontech, Mountain View, CA)] was from a single donor. In these 24 kinds of human total RNA, 23 RNAs excluding skeletal muscle were used to determine expression localization of 15 human UGT isoforms. Donor information for these samples is shown in Table 1. Total RNA was reverse-transcribed by the method described above.
Results
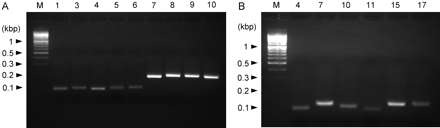
Specificity of Quantification. Detection of 15 kinds of human UGTs was achieved with the specific primers shown in Table 2. All amplicon sequences were identical with each subtype sequence when the specific primer pair was used to prepare the vector for a calibrator of the UGT1A subfamily. However, amplicon length was too long for the real-time PCR quantification system, so the primers of the UGT1A subfamily were redesigned. The real-time PCR reaction was carried out using the human esophagus cDNA library for UGT1A7, human small intestine cDNA for UGT1A8 and UGT1A10, and human liver cDNA for all other subtypes. In all cases, fluorescence with SYBR Green incorporation was at least detected (data not shown). To verify the specificity of the PCR reaction, the reaction mixture was applied to an agarose gel and electrophoresed. Even after 45 PCR cycles, single bands with the expected sizes were detected in all reactions on a 2.5% agarose gel after ethidium bromide staining (Fig. 1). All bands were cut out and sequenced, and the identity of all predicted sequences was confirmed (data not shown). From these results, the fractional determination of 15 kinds of human UGTs (1A1, 1A3, 1A4, 1A5, 1A6, 1A7, 1A8, 1A9, 1A10, 2B4, 2B7, 2B10, 2B11, 2B15, and 2B17) was confirmed at the mRNA level by real-time PCR using the specific primers.
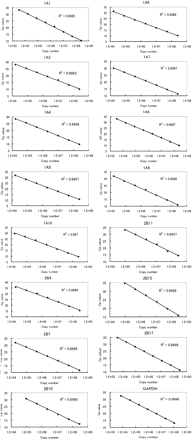
Calibration Curve. Quantification of mRNA expression of human UGT isoforms and human GAPDH was performed by reading the copy number of each sample from the crossing point (Cp) value on individual calibration curves. The calibration curves were prepared from the Cp values of serial diluted calibrator vectors of individual UGTs or GAPDH as the real-time PCR template with each specific primer pair. For the 15 kinds of human UGT and human GAPDH, all calibration curves showed linearity between Cp value and indicated copy number with correlation coefficients greater than 0.9957 and high repeatability, indicating a precise log-linear relationship (Fig. 2). For each target, the area obtained by the linearity represented the quantitative range and the area displayed as the standard curve at the region of low copy number represented the quantitative limit.
Determination of Human UGTs and Localization in Various Human Tissues. The exhaustive measurement method established here for human UGT enzymes at the transcriptional level was applied to the comparison of the UGT expression in a total of 23 kinds of human tissue. The quantification data are defined as the copy numbers of UGT transcripts normalized to GAPDH. Although GAPDH expression was slightly high in tissues, such as the testes, heart, brain, and cerebellum, almost equal copy numbers were observed in most human total RNA samples (Tables 3 and 4). Most of the UGT family enzymes were expressed in the liver and alimentary system, especially UGT2B7, an enzyme that is known to be expressed in the liver, small intestine, and colon, as well as the kidney (Parquet et al., 1985; Girard et al., 2003). The results shown in Tables 3 and 4 and Fig. 3 confirm that most of the human UGT family enzymes are expressed in the alimentary system and liver.
Results of quantitative determination for the UGT1A subfamily
Values shown are copy number × 104 normalized with GAPDH, mean ± S.D., n = 3 to 4. Donor information is presented in Table 1.
Results of quantitative determination for the UGT2B subfamily
Values shown are copy number × 104 normalized with GAPDH, mean ± S.D., n = 3 to 4. Donor information is presented in Table 1.
Agarose gel electrophoresis of PCR products showing specificity of real-time RT-PCR for human UGT1A subfamily isoforms (A) and UGT2B subfamily isoforms (B). A human cDNA library, which were prepared by the RT reaction from human esophagus total RNA for 1A7, human small intestine total RNA for 1A8 and 1A10, and human liver total RNA for all other UGTs, was used as PCR templates, and real-time PCR using specific primers was performed as described under Materials and Methods. Electrophoresis was performed on 2.5% agarose gel at 100 V in 1× Tris borate-EDTA buffer. Lane M, 100 bp. DNA ladder marker (M) and isozyme numbers for each subfamily are indicated at the top of each gel. Expected amplicon lengths were as follows: 1A1, 96 bp; 1A3, 100 bp; 1A4, 96 bp; 1A5, 99 bp; 1A6, 105 bp; 1A7, 186 bp; 1A8, 198 bp; 1A9, 200 bp; 1A10, 198 bp; 2B4, 104 bp; 2B7, 123 bp; 2B10, 111 bp; 2B11, 96 bp; 2B15, 124 bp; and 2B17, 124 bp. Arrowheads indicate DNA size in kilobase pairs (kbp).
Of all UGT isoforms in the 23 human tissues examined, the most abundantly expressed UGT isoforms in the total RNA preparation of each human tissue were UGT2B4 (liver) followed by UGT2B15 (liver), which, respectively, had 8.98 and 4.38 times greater expression than UGT2B7 (liver). The level of UGT2B10 (liver) was identical to that of UGT2B7, yet the expression of UGT2B10 was specific to the liver, unlike that of 2B7. The expression level of UGT1A1 was approximately 30% of the UGT2B7 expression and was almost the same level as UGT1A9 (liver).
UGT2B4 and UGT2B7 are, reportedly, expressed in the kidney as well as in the liver, and UGT2B7 is responsible for glucuronidation of endogenous steroids such as gluco- and mineralocorticoids in the kidney (Girard et al., 2003). UGT2B7 was, indeed, expressed in the kidney; however, the UGT2B4 expression level in the kidney was much lower than that of UGT2B7, and the most abundantly expressed UGT in the kidney was UGT1A9, with a level 124% of UGT2B7 expression. Moreover, UGT1A9 expression in the kidney was 4.9 times higher than that in the liver. UGT1A6 was also expressed in the kidney, but the level was approximately 13.4% that of UGT1A9.
Calibration curves for 15 human UGT isoforms and human GAPDH quantification by real-time PCR. Each calibrator vector (1 ng/μl for all UGT isoforms or 0.5 ng/μl for GAPDH) was prepared with a series of 5-fold serial dilutions in sterilized water to ×9,765,625 dilution. Aliquots (5 μl) of diluted vectors were subjected to real-time PCR under the condition described under Materials and Methods using the respective specific primers listed in Table 1. The linear area of copy numbers against the Cp value in each calibration curve was regarded as the respective quantification area. Each point represents the mean (n = 4) of duplicate data from two separate analyses.
In the small intestine and colon, several UGT isoforms, such as UGT1A1, 1A10, 2B7, 2B15, and 2B17, were expressed. UGT1A10 and UGT2B17 were found predominantly in the intestines. In the esophagus, UGT1A5, 1A7, 1A9, 1A10, and 2B4 were expressed, and the most abundantly expressed UGT isoform was 1A7; however, all levels of expression were relatively low. In addition, even though expression levels were very low, the steroidogenic tissues, such as the breast, prostate, heart, and adrenal gland, expressed several kinds of UGT isoforms.
Discussion
The isoforms of the UGT1 family are all transcribed from a single gene, located on chromosome 2q37, by alternate splicing of four constant exons (exons 2–5), which encode for a carboxyl terminal moiety, to a variable exon 1 encoding an amino terminal moiety for each protein (Zhang et al., 2004). In contrast, the isoforms of the UGT2 family are products of separate genes clustered on chromosome 4q13 and are divided into the UGT2A and UGT2B subfamilies (Mackenzie et al., 1997). The real-time PCR methods including each isoform-selective PCR primer set designed for 15 UGT isoforms have high isoform specificity, and the expression level of UGT isoform transcripts (excluding UGT2A1 and UGT2B28, which have not yet been detected) has been determined in the various human tissues. Measurement results are shown in detail in Tables 3 and 4 and summarized in Fig. 3. From the quantitative determination of UGT isoforms, high expression is predominant in the liver, gastrointestinal tract (small intestine and colon), and kidney.
Determination of 15 human UGT isoforms and localization in 23 human tissues shown in Tables 3 and 4. Twenty-three kinds of human cDNA were diluted 6 times with sterilized water, and 5 μl of each sample was subjected to real-time PCR under the condition described under Materials and Methods using the respective specific primers listed in Table 2. The units of the y-axis represent the UGT copy number × 104 normalized to GAPDH, and the 37,900 and 18,500 values are included as off-scale data of UGT2B4 and UGT2B15, respectively. Each data point represents the mean value (n = 3–4), and error bars were omitted for clearer viewing. All of the data were reconfirmed by repeated examination.
In the liver, many isoforms, such as UGT1A1, 1A3, 1A4, 1A5, 1A6, 1A8, 1A9, 2B4, 2B7, 2B10, 2B15, and 2B17, are reported to be expressed (Strassburg et al., 1997b, 2000; Tukey and Strassburg, 2001). Here, UGT1A1, 1A3, 1A4, 1A5, 1A6, 1A7, 1A9, 2B4, 2B7, 2B10, 2B15, and 2B17 have been localized. We were surprised to find that quantitative reverse transcription real-time PCR reveals the extremely high expression of UGT2B4 and UGT2B15. UGT2B4, which is known as a bile acid glucuronidating enzyme, catalyzes the glucuronide conjugation with various molecules, including bile acids, 5α-reduced androgens, catecholestrogens (17-epiestriol and 4-hydroxyestrone), and phenolic and monoterpenoid compounds (Fournel-Gigleux et al., 1989; Ritter et al., 1992; Pillot et al., 1993; Lévesque et al., 1999; King et al., 2000; Finel et al., 2005). Moreover, UGT2B4 is well known to be exclusively expressed in human liver and not in kidney (Pillot et al., 1993) or expressed in both liver and kidney (Girard et al., 2003). The highest expression level of UGT2B4 is quantitatively confirmed to be in the liver, whereas UGT2B4 is expressed in very low levels in heart, testis, kidney, thymus, trachea, esophagus, and placenta. This isoform seems to be the most abundant UGT2B isoform in the liver. UGT2B15 is abundantly detected in the liver, and also detected in the stomach, breast, small intestine, and colon, as well as prostate in relatively large amounts. UGT2B15 has been fully characterized for its ability to conjugate with drugs, environmental pollutants, and dietary components (Green et al., 1994) and shares more than 95% homology with UGT2B17, which is one of the most important conjugate enzymes in androgen metabolism (Turgeon et al., 2003).
Our data from this quantitative determination of human UGT isoform transcripts are generally consistent with those of previous individual reports. For example, UGT1A8 is expressed in the intestine and colon but not in the liver (Cheng et al., 1998; Tukey and Strassburg, 2000). We also determined UGT1A8 expression in the small intestine and colon and in the bladder and trachea. UGT1A9, an estrogen conjugation enzyme, which contributes to the glucuronidation of 4-hydroxyestradiol and 4-hydroxyestrone, is known to be abundantly expressed in the kidney, as well as in the small intestine, ovary, and testis at low levels (Albert et al., 1999). We detected the highest expression in the kidney followed by the liver, and considerable expression in the adrenal glands, small intestine, and colon. UGT2B7 is able to glucuronidate various steroid hormones (such as androsterone and epitestosterone) and fatty acids (Radominska-Pandya et al., 2001) and is able to conjugate with many drug, such as morphine, ketoprofen, and all-trans retinoic acids. We found that this isoform is expressed in the liver, small intestine, and colon, as well as in the kidney; these results are identical with previous reports (Parquet et al., 1985; Girard et al., 2003). UGT2B10 has been known to be an orphan isoform that has been reported recently to play a major role in nicotine inactivation by direct conjugation with glucuronic acid at the aromatic nitrogen atom and to express in the liver (Kaivosaari et al., 2007). We also found that the 2B10 isoform is detected in the liver and not in other various tissues. UGT2B17 is able to conjugate with C-19 steroids, dihydrotestosterone, androstane-3α,17β-diol, testosterone, and androsterone, and its expression has been found in various tissues, including the liver, kidney, testis, uterus, placenta, mammary gland, adrenal glands, skin, and prostate by Southern blot analysis (Beaulieu et al., 1996). We detected UGT2B17 in the small intestine and colon, as well as in other various tissues, including considerable amounts in the liver. However, some differences in expression of UGT2B17 were also found. Kaivosaari et al. (2007) reported that UGT1A4 was detected in trachea, kidney, small intestine, and colon at comparatively low levels relative to those in the liver using real-time PCR, whereas we only detected it in liver. Moreover, Collier et al. (2002) reported that most UGT2B isoforms were expressed in human placenta using RT-PCR, whereas we detected only UGT1A5 in the placenta. The discrepancy in these results may be due to 1) the high interindividual variability in UGT isoform expression levels and thus the necessity for an exhaustive quantitative determination method and 2) configuration of a slightly lower amplification efficiency with use of the quantification primer set, such as UGT2B10, UGT2B11, and UGT2B15 (Fig. 2). The possibility of variability in the expression level is confirmed by previous reports of interindividual differences in expression for UGT1A1 (Aueviriyavit et al., 2007) and UGT1A5 (Finel et al., 2005). The possibility of low amplification efficiency is supported by our detection of UGT2B11 in the liver (Fig. 1) with the expected sequence, but without levels of amplification that correlated well between the copy number and Cp value. Therefore, these results failed to provide a precise quantitative determination beyond the limit of detection (Fig. 3; Tables 3 and 4), although significant levels of UGT2B11 expression were confirmed in the human hepatic carcinoma cell line (HepG2) (unpublished data). Our major objective was to construct a speedy and multiple determination method of human UGTs, including the ability to directly compare expression levels among different UGT isoforms using real-time PCR; thus, this method is significantly different from simple RT-PCR detection of UGT isoforms.
The range of human samples analyzed in this experiment is limited by the commercial availability and number of the analytes. Of the 23 human total RNA samples analyzed by real-time RT-PCR, 4 total RNA samples from the stomach, cerebellum, breast, and adrenal gland are derived from a single donor, and, therefore, the data have been recorded as a pro forma value. The other 19 samples are pooled total RNA from three donors; therefore, n = 3, which means the same determination is repeated three times. The UGT expression levels obtained reflect the average levels from three people and the errors due to measurement. The quantitative data in this experiment indicate the copy number of transcripts per unit total RNA preparation of each tissue. Therefore, the comparison of total expression levels for one UGT between different tissues is difficult; however, the data are applicable when the expression levels between different UGT isoforms in the same tissue are compared. The total RNA samples are reflective of the tissue as a whole. Further studies with other total RNA samples are required if colocalization data in the same tissue are needed. In addition, further studies are needed to construct the complete quantification system of human UGTs, including UGT2A1, UGT2B28, and variants of some UGT isoforms. It should also be mentioned that while this manuscript was being prepared, one study reported the expression profiles of all functional UGT1A and UGT2B isoforms in 12 kinds of normal human tissues using RT-PCR (Nakamura et al., 2008). This new study detected UGT2B11 expression at considerable levels in liver, bladder, and breast, although UGT2B11 expression was not detected quantitatively in any of the 23 human total RNA samples here. To explain these different results in detail, further investigation may be needed for UGT2B11 determination.
Recently, the serious adverse effect of irinotecan has been closely correlated to the transcriptional activity of UGT1A1, and related clinical trials with individual patients are being investigated. In the future, our simple and precise real-time RT-PCR quantification method for all quantification of UGT isoforms using an identical annealing temperature may provide valuable information about the medical efficacy or pharmacokinetic characteristics of a wide variety of drugs that are metabolized by UGTs as this method is very useful for determining human UGT transcripts owing to its high isoform selectivity and specificity. It is hoped that this method will be more broadly available for the determination of human UGT isoforms.
Acknowledgments
We express our gratitude to Dr. Tsutomu Suzuki (Department of Toxicology, Hoshi University School of Pharmacy and Pharmaceutical Sciences) for valuable advice and Satoe Arai for excellent technical assistance.
Footnotes
-
This work was supported in part by a grant-in-aid from the Ministry of Education, Culture, Sport, Science and Technology of Japan.
-
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
-
doi:10.1124/dmd.108.023598.
-
ABBREVIATIONS: UGT, UDP-glucuronosyltransferase; RT, reverse transcription; PCR, polymerase chain reaction; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; bp, base pair; Cp, crossing point.
- Received July 28, 2008.
- Accepted October 3, 2008.
- The American Society for Pharmacology and Experimental Therapeutics