Abstract
Microsomal protein per gram of liver (MPPGL) is an important scaling factor for bottom-up physiology-based pharmacokinetic modeling and simulation, but data in pediatrics are limited. Therefore, MPPGL was determined in 160 liver samples from pediatric (n = 129) and adult (n = 31) donors obtained from four sources: the University of Maryland Brain and Tissue Bank (UMBTB), tissue retrieval services at the University of Minnesota and University of Pittsburgh, and Sekisui-Xenotech. Tissues were homogenized and subjected to differential centrifugation to prepare microsomes, and cytochrome c reductase activities in tissue homogenates and microsomes were used to estimate cytochrome P450 reductase (POR) activity as a marker of microsomal recovery; microsomal POR content was also assessed by quantitative proteomics. MPPGL values varied 5- to 10-fold within various age groups/developmental stages, and tissue source was identified as a contributing factor. Using a “trimmed” dataset comprised of samples ranging from 3 to 18 years of age common to the four sources, POR protein abundance and activity in microsomes and POR activity in homogenates was lower in UMBTB samples (autopsy) compared with other sources (perfused/flash-frozen). Regression analyses revealed that the UMBTB samples were driving an apparent age effect as no effect of age on log-transformed MPPGL values was observed when the UMBTB samples were excluded. We conclude that a mean±SD MPPGL value of 30.4±1.7 mg/g is representative between one month postnatal age and early adulthood. Potential source effects should be considered for studies involving tissue samples from multiple sources with different procurement and processing procedures.
SIGNIFICANCE STATEMENT Microsomal protein per gram of liver (MPPGL) is an important scaling factor for bottom up PBPK modeling and simulation, but data in pediatrics are limited. Although MPPGL varies 5- to 10-fold at a given developmental stage, a value of 30.4 ± 1.7 mg/g (mean ± SD) is representative between one month postnatal age and early adulthood. However, when tissue samples are obtained from multiple sources, different procurement and processing procedures may influence the results and should be taken into consideration.
Introduction
The value of pharmacokinetic (PK) modeling and simulation in the design and conduct of pediatric clinical trials is now well recognized by the pharmaceutical industry and is also routinely employed by regulatory agencies tasked with evaluating data presented in support of pediatric labeling (Germovsek et al., 2019). Clinically, PK modeling and simulation plays an important role in describing drug disposition and response on a population basis, and the identification of factors that account for observed inter-individual variability in pediatric or other patient populations can be leveraged to develop models to individualize treatment (Krekels et al., 2017; Neely et al., 2018). “Top down” models, consisting of a limited number of empirical equations, describe the distribution of data observed for a given drug in a defined population, but the ability to extrapolate from one drug to another in the population of interest, or from one patient population to another, is limited. On the other hand, “bottom up” physiology-based pharmacokinetic (PBPK) models combine drug-specific and system-specific information, allowing a variety of physiochemical, in vitro preclinical and in vivo clinical data to be integrated, allowing reproduction of the underlying anatomy and physiology of the biologic system (Maharaj and Edginton, 2014).
Given the relative abundance of data generated in adults or adult-derived systems, multiple strategies have been applied to scale values for parameters, such as plasma clearance in vivo or intrinsic clearance of unbound drug in vitro, from adults to pediatric patients of various ages and developmental stages. Allometric scaling solely on the basis of weight has limitations, especially in newborns, infants, and young children (Edginton and Willmann, 2006; Mahmood et al., 2014; Calvier et al., 2017); these limitations have been addressed by adding maturation functions that characterize the developmental trajectories of drug-metabolizing enzymes contributing to drug clearance (Calvier et al., 2019).
“Bottom up” PBPK models require additional scaling factors—one that addresses the changes in liver volume that accompany growth and development, and one that scales in vitro intrinsic clearance of unbound drug to hepatic clearance (CLH). Johnson et al. (Johnson et al., 2005) compiled a dataset of 5,036 liver volume measurements from nine different published studies, and after assessment of several potential covariates, determined that a model expressing liver volume as a function of body surface area (liver volume = 0.722*BSA1.176) best described the change in liver volume between birth and 18 years of age. The group also found that the model predicted liver volume in adults with precision and accuracy that exceed almost all (10/11) published adult models (Johnson et al., 2005).
Data characterizing the second scaling factor, microsomal protein per gram of liver (MPPGL), are much more limited, especially across the pediatric age range. In the largest study to date (n = 128), MPPGL values from adult samples aged 20 to 75 years of age were not normally distributed and varied 19-fold from 6.7 to 128.0 mg/g liver, with a mean ± SD of 39.5 ± 21.6 mg/g liver (Zhang et al., 2015). This mean MPPGL value of 40 mg/g is in agreement with the value presented by Hakooz et al. (Hakooz et al., 2006) as well as the conclusion of a meta-analysis of several previously published studies that includes the Hakooz study (Barter et al., 2007; Barter et al., 2008). However, in this analysis, samples covering the period between birth and adulthood (18 years of age) were limited as only one pediatric sample (11 years of age) was reportedly included in the original meta-analysis (Barter et al., 2007); in a follow-up study, the meta-analysis dataset was supplemented with four additional pediatric donors aged 2, 4, 9, and 13 years of age (Barter et al., 2008). Thus, available data regarding the developmental trajectory of MPPGL include only five time points between 2 and 13 years of age and are the basis of age-dependent MPPGL estimates used in pediatric PBPK models, such as the value of 26 mg/g used in a recent PBPK modeling and simulation study in infants (Salerno et al., 2021). Therefore, the purpose of this investigation was to address the paucity of data describing the ontogeny of MPPGL between birth and adulthood given their importance as scaling factors for PBPK modeling and simulation.
Materials and Methods
Materials and Reagents.
β−NADPH and cytochrome c derived from horse heart were purchased from Sigma Aldrich Chemical Co. (St. Louis, MO, USA). Sodium dithionite was obtained from Merck (Damstadt, Germany). Pooled human liver microsomes (n = 200 donors, mixed gender, 100 male and 100 female, Lot No. 1410230) and pooled human liver homogenate (n = 20 donors, mixed gender, Lot No. 1510072) were purchased from Xenotech, LLC, (Lenexa, KS, USA) and used as controls in the cytochrome c reductase assay. All other chemicals were of reagent grade and were purchased from either Sigma Aldrich Chemical Co. or Thermo-Fisher Scientific (Fairlawn, NJ).
Liver Samples.
A total of 160 liver samples (n = 129 pediatric and n = 31 adult) were included in this study. The primary sources of tissue were the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)-supported tissue retrieval program at the Brain and Tissue Bank for Developmental Disorders at the University of Maryland (UMBTB, Baltimore, MD (now the University of Maryland Brain and Tissue Bank, and a member of the NIH NeuroBioBank network); n = 60, including n = 52 pediatric, and n = 8 adult samples) and the NIH-supported Liver Tissue Cell Distribution System (LTCDS), with sites at the University of Minnesota (n = 34 pediatric) and the University of Pittsburgh (n = 37, comprised of n = 14 pediatric and n = 23 adult samples). Additional pediatric liver samples were obtained from In Vitron (Tucson, AZ; n = 4), the University of Miami (n = 1), and the Association of Human Tissue Users (n = 1). In addition, homogenates and microsomes isolated from pediatric livers for this study (n = 23) were donated by Xenotech, LLC. (Lenexa, KS; now Sekisui Xenotech, Kansas City, KS). The distribution of samples by tissue source and age group is provided in Supplemental Table 1, and available demographic data, including reported cause of death (when available), for all samples is listed in Supplemental Table 2. Overall, postnatal samples ranged in age from birth to 79 years of age; 57/160 (35.6%) were female, 102 (63.8%) were male, and sex was unknown for one sample. Race was reported as African American for 40 samples (25.0%), Caucasian for 76 samples (47.5%), Hispanic for 7 samples (4.4%), and Native American and Pacific Islander for one sample each; race information was not available for 35 samples (21.9%), including all samples from the University of Minnesota (n = 34). Use of the tissue samples was classified as non-human subjects research by the Children’s Mercy Pediatric Institutional Review Board. Tissues were stored at -70°C or below prior to preparation of subcellular fractions.
Assessment of RNA Integrity.
As a surrogate measure of liver tissue quality for MPPGL determination, RNA integrity was assessed at the time of tissue processing. Briefly, total RNA was extracted from an average of 35 mg of human liver tissue using a Qiagen RNAeasy kit (Qiagen, Valencia, CA) following the manufacturer’s protocol, including the on-column DNase I digestion step. The quantity of isolated RNA was assessed using a NanoDrop 1000 instrument which informed the amount of sample for subsequent analysis on an Experion StdSens RNA microfluidic chip (Bio-Rad, Hercules, CA; cat #700-7103). This Experion RNA analysis allowed us to determine total RNA and mRNA integrity, purity, and concentration. Specifically, the generated electropherogram enabled evaluation of the RNA sample for degradation. The RNA quality index (RQI) is calculated by the Experion software, with an RQI of 1 representing highly degraded and an RQI of 10 representing high-quality total RNA. Further details on the method, representative examples and the development and validation of RQI as an RNA quality indicator can be obtained from Bio-Rad technical note 5761 available at https://www.gene-quantification.de/Bio-Rad-bulletin-5761.pdf.
Preparation of Pediatric Liver Homogenates and Microsomes.
Human liver microsomes were prepared by differential centrifugation, essentially as described by Lu and Levin (Lu and Levin, 1972). Briefly, pre-weighed, frozen liver samples were placed in homogenizing buffer (∼3 mL/g liver; 50 mM Tris.HCl, pH 7.4 at 4°C, containing 150 mM KCl and 2 mM EDTA) and allowed to thaw at 4°C. Liver samples were quickly minced with dissecting scissors on ice, placed in Potter-Elvehjem-type glass mortars (round-bottom) and homogenized on ice with a Polytron tissue homogenizer (Kinematica USA, Bohemia, NY, USA) using 3–4 second bursts of grinding for 1–2 passes. Liver samples were subjected to further homogenization (3–4 strokes on ice, 2–3 passes) in the glass mortars with Teflon pestles utilizing a motor-driven tissue homogenizer (Caframo Model BDC-3030, Wiarton, ON, Canada). Homogenates were placed into low-speed centrifuge tubes, filled with homogenization buffer, briefly mixed, an aliquot of homogenate removed, and the volume recorded. Subsequently, nuclei and lysosomes were removed from the homogenate by centrifugation (800 gmax for 15 minutes at 4°C). The resulting supernatant was further centrifuged (12,000 gmax for 20 minutes at 4°C), and the supernatant fraction was subjected to ultra-centrifugation (105,000 gmax for 70 minutes at 4°C). The resulting supernatant (cytosol) was stored at -80°C for future determination of cytosolic protein per gram liver ontogeny. The pellet (microsomal fraction) was removed from the centrifuge tube, transferred to a low-volume glass mortar, manually re-suspended in 0.25 M of sucrose with a Teflon pestle, and stored at -80°C until use. Protein concentrations were determined with a Micro BCA Protein Assay kit (Pierce Chemical Co., Rockford, IL, USA) using bovine serum albumin (Sigma, St. Louis, MO, USA) as the standard.
Cytochrome C Reductase (POR) Activity.
In vitro cytochrome P450 reductase (POR) activity was used as the marker for microsomal content and enzyme activity assays were performed in 96-well microtiter plates. Incubations (200 μl) contained human liver homogenate (typically 30 μg of homogenate protein, range 9.5-65 μg), or microsomes (typically 3 μg of microsomal protein, range 3-21 μg), potassium phosphate buffer (350 mM, pH 7.4), MgCl2 (3 mM), EDTA (1 mM), and cytochrome c (50 μM) at the final concentrations listed. Although homogenate studies were conducted in a matrix with higher protein concentrations relative to the microsomal studies, the concentration of cytochrome c present in the performed incubations is expected to be above enzyme saturating concentrations such that free, unbound concentrations of substrate are comparable in homogenates and microsomes, should higher non-specific binding be present in the latter. Each microtiter plate contained oxidized and reduced cytochrome c standards created by adding 50 μl of water or sodium dithionite (250 mg/mL), respectively, to each standard-containing well for a total volume of 250 μl. Plates were pre-incubated at 30±1°C for 5 minutes directly in a Bio-Tek Synergy HT multi-mode micro-titer plate reader, and reactions were initiated in sample wells by the addition of 50 μl of β−NADPH (500 μM). Absorbance (550 nm) was recorded with the plate reader operating in kinetic mode at 25 second scan intervals for 5 minutes. POR activity was defined as the rate of reduction of cytochrome c, determined by measuring the rate of change in optical density (OD) in the linear portion of the kinetic curves and calculated using Eq. 1:
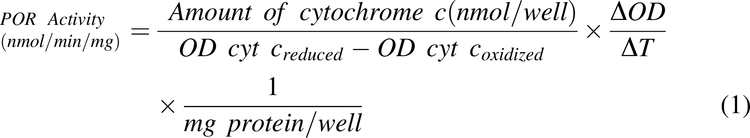 where OD cyt coxidized is the mean of optical densities from wells containing oxidized cytochrome c, OD cyt creduced is the mean of optical densities from wells containing reduced cytochrome c and ΔOD/ΔT is the mean change in OD during the linear portion of the curve.
where OD cyt coxidized is the mean of optical densities from wells containing oxidized cytochrome c, OD cyt creduced is the mean of optical densities from wells containing reduced cytochrome c and ΔOD/ΔT is the mean change in OD during the linear portion of the curve.
Estimation of Liver Microsomal Protein Content.
The total amount of homogenate protein obtained per gram of starting liver tissue was calculated as:
 where ProteinHom and VolumeHom represent the total amount and volume of homogenate prepared from the starting amount of liver tissue, respectively.
where ProteinHom and VolumeHom represent the total amount and volume of homogenate prepared from the starting amount of liver tissue, respectively.
MPPGL was corrected for recovery of microsomal protein during the preparation procedure using the following equation (Wilson et al., 2003; Barter et al., 2008):
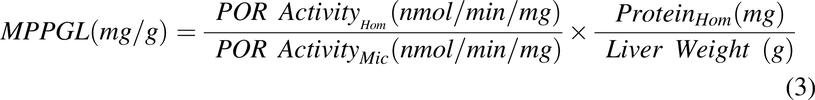 where POR ActivityHom and POR ActivityMic represent the rate of reduction of cytochrome c in the cellular homogenate and microsomal fractions, respectively.
where POR ActivityHom and POR ActivityMic represent the rate of reduction of cytochrome c in the cellular homogenate and microsomal fractions, respectively.
Recovery of microsomal protein from liver homogenate was determined as:
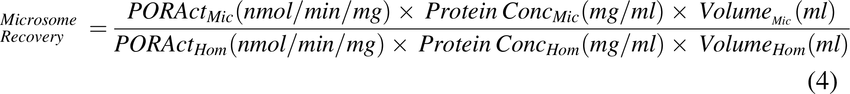
Assessment of POR Content by Quantitative Proteomics.
POR content was determined in a subset of samples (n = 123) by a surrogate peptide-based liquid chromatography with tandem mass spectroscopy (LC-MS/MS) method (Bhatt and Prasad, 2018). The surrogate peptide of POR quantification (Supplemental Table 3) was selected based on previously reported criteria (Bhatt and Prasad, 2018) and was obtained from New England Peptides (Boston, MA). A previously optimized protocol for trypsin digestion of microsomal proteins and the sample preparation for liquid chromatography with tandem mass spectroscopy analysis was adopted (Bhatt et al., 2018). Briefly, 60 μl of the human liver microsome (2 mg/ml) sample was denatured and reduced by incubation with 40 μl of ammonium bicarbonate digestion buffer (100 mM, pH 7.8) and 10 μl of 100 mM dithiothreitol at 90°C for 5 minutes. The resultant solution was alkylated by adding 20 μl of 200 mM iodoacetamide at room temperature for 20 minutes. The processed protein was then precipitated using ice-cold methanol (500 μl) and chloroform (200 μl). Water (400 μl) was added to the mixture, and the sample was vortexed and centrifuged at 12,000 x g for 5 minutes. The upper solvent layer was carefully removed, and the protein pellet was washed with 500 μl of ice-cold methanol followed by centrifugation at 12,000 x g for 5 minutes. The final protein pellet was dissolved in 60 μl of ammonium bicarbonate. The samples were then digested by trypsin (protein:trypsin ratio of ∼50:1) in a final volume of 80 μl at 37°C for 16 hours. The reaction was stopped by adding 20 μl of peptide internal standard (prepared in 70% acetonitrile in water containing 0.1% formic acid) and 10 μl of the neat solvent (70% acetonitrile in water containing 0.1% formic acid). The samples were centrifuged at 4000 x g for 5 minutes. The calibration curve standards were prepared by spiking standard working solutions of peptides (prepared in 70% acetonitrile in water containing 0.1% formic acid) into ammonium bicarbonate buffer. Eight calibration standards were used to produce a range of on column amounts from 0.3 to 150 fmol. For each standard, working stock solutions of the peptide (10 μl) were added in the last step instead of the neat solvent. All of the human liver microsome samples were digested and processed in triplicate.
Protein quantification was performed using a triple-quadrupole MS instrument (Sciex Triple Quad 6500, Concord, ON) in ESI positive ionization mode coupled to an Acquity UPLC, I-class (Waters, Milford, MA). Five μl of each trypsin digested sample was injected onto the column (ACQUITY UPLC HSS T3 1.8 μm, C18 100A; 100 × 2.1 mm, Waters, Milford, MA). Surrogate light and heavy (internal standards) peptides were monitored using instrument parameters provided in Supplemental Table 3. The LC-MS/MS data were processed using Analyst 1.6.2 version software (Sciex, Concord, Ontario). The lower limit of quantification (LLOQ) of POR peptide was 0.07 pmol/mg protein. The protein expression data are reported as the mean and standard deviation (SD) of values obtained in at least three experiments.
Data and Statistical Analyses.
The distribution of raw and log-transformed MPPGL values was assessed for normality by visual inspection of the normal quantile plot and by the Shapiro Wilk W test (Supplemental Fig. 1). To characterize the relationship between log-transformed MPPGL (log MPPGL) and developmental stage, liver samples were stratified and characterized as recommended by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (Williams et al., 2012). Specifically, samples were categorized as neonatal (birth to 28 days; n = 3), infant (29 days to 12 months; n = 17), toddler (13 months to 2 years; n = 9), early childhood (2 to 5 years; n = 21), middle childhood (6 to 11 years; n = 32), early adolescence (12-18 years; n = 47), and late adolescence (19-21 years; n = 0). Adult samples were arbitrarily divided into two additional categories of younger adult (21 to 50 years; n = 16) and older adult (> 50 years; n = 15).
A subset of the data were used to assess the potential influence of factors related to tissue source. The data subset for this analysis, hereafter referred to as the “trimmed” dataset, comprised samples from an age range, 3 to 18 years, that was common to the major sources of samples: UMBTB, LTCDS sites at Minnesota and Pittsburgh and Sekisui-Xenotech. Comparisons of the tissue sources were conducted using analysis of variance (ANOVA) followed by post-hoc analysis using Tukey’s Honestly Significant Difference, or Welch’s ANOVA in the presence of unequal variance. In addition, a linear model was fit to the trimmed data set, modeling log(MPPGL) as a function of sample source, sex, PORHom value, age, and age (centered)2. The resulting source effects were taken as estimates of the effect of source after controlling for any effects of sex, PORHom value, and age. Finally, each log(MPPGL) value was adjusted in the full sample by subtracting the appropriate source effect estimate. The effectiveness of the adjustment was checked by refitting the model to the source-adjusted log(MPPGL) values for the trimmed sample and verifying that all source effects were zero. The remaining analyses were carried out using these source-adjusted log(MPPGL) values. Because source effects could not be assessed for the single sample sources (Miami and Association of Human Tissue Users samples), their log(MPPGL) values were not adjusted.
Comparisons across age strata were conducted by ANOVA followed by post-hoc analysis using Tukey’s Honestly Significant Difference test. All statistical analyses were conducted in JMP Pro version 14.2.0, SAS version 9.4 (SAS Institute Inc., Cary, NC), and R (R Core Team, Vienna, Austria). The relationship between source-adjusted logMPPGL and age as a continuous variable was also assessed using GraphPad Prism v8 (GraphPad Software, San Diego, CA) and an asymptotic exponential or sigmoid Emax with Hill coefficient models (Anderson and Holford, 2008) to characterize the developmental trajectory up to age 18 for use in PBPK applications. Output for the source-adjusted MPPGL model was compared with values up to age 18 years generated using the polynomial function reported by Barter et al. (2008):
MPPGL (mg/g) = 10^(1.047 + 0.01579age - 0.000382age2 + 0.00000237age3
Results
General Distribution of MPPGL Values.
Overall, postnatal MPPGL values varied 10-fold (7.9 mg/g to 80.8 mg/g) across the entire age range of birth to 79 years and were not normally distributed based on visual inspection of normal quantile plots and the Shapiro Wilk W test (P < 0.0001). Log-transformation of the MPPGL data resulted in normal distributions according to the same criteria (output of the JMP Pro analysis can be found in Supplemental Fig. 1).
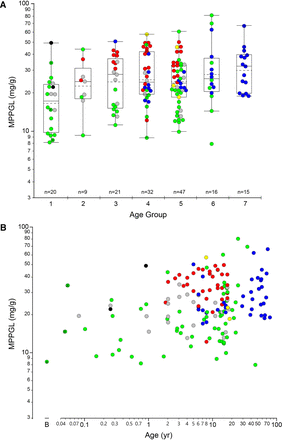
The geometric mean for MPPGL in all samples (n = 160) was 24.0 mg/g, with a median value of 24.1 mg/g and an interquartile range of 16.6 mg/g to 34.3 mg/g. The distribution of logMPPGL values as a function of age group/developmental stage is presented in Fig. 1A and with age as a continuous variable in Fig. 1B, individual samples and color-coded by tissue source. There were only three samples within the neonatal age range, so they have been combined with the “infant” group and are designated with a white “x” within the corresponding symbol. A summary of the distribution of MPPGL values in each age group/developmental stage is also provided in Table 1.
Relationship between log-transformed MPPGL values and postnatal age. Panel A. Postnatal age is expressed as a categorical variable using NIH-recommended age strata (Williams et al., 2012): Group 1, Infancy (28 days to 12 months of age); Group 2, Toddler (13 months to 2 years of age); Group 3, Early Childhood (2 years to 5 years of age); Group 4, Middle Childhood, (6 years to 11 years of age); Group 5, Early Adolescence (12 years to 18 years of age). For reference, Groups 6 and 7 represent Younger Adults (19 years to 50 years of age) and Older Adults (> 50 years of age), using an age of 50 years as an arbitrary cut-off. Box plots were constructed using the “outlier” format in JMP Pro 14.3. Boxes are defined by the first and third quartiles (25th and 75th quantiles, respectively), and the median is indicated by the horizontal line within the boxes; the sample mean is indicated by the dashed horizontal line. The whiskers extend from the ends of the box to the outermost data point that falls within the distance calculated as the third quartile + 1.5*(interquartile range) at the upper bound and the first quartile - 1.5*(interquartile range) at the lower bound; points extending beyond the whiskers are considered outliers. Panel B. Age is presented as a continuous variable, “B” indicates day of “birth”. For both panels, data points are colored according to source of tissues: UMBTB (green), University of Minnesota (red), University of Pittsburgh (blue), Sekisui-Xenotech (gray), Vitron (yellow) and other (University of Miami and Association of Human Tissue Users, one sample each; black). The three points with a black “x” in the center (all from UMBTB; green) represent neonatal samples from the day of birth to 28 days postnatal age.
Distribution of MPPGL values within each age group or developmental stage according to NICHD-recommended age strata
The dataset included only three neonatal samples (birth to 28 days postnatal age) that were included in the “Infant” group.
Variability in MPPGL Values: Contribution of Tissue Source.
Inspection of the data presented in Fig. 1A and Table 1 reveals that MPPGL values varied 5- to 10-fold within various age groups/developmental stages. Visual inspection of the data color-coded by source of liver tissue further reveals that several age groups include samples from multiple sources, whereas others (e.g., Group 7, older adults) include samples from a single source. This observation suggests the possibility that source of tissue may confound interpretation of any effect of age on MPPGL values. To assess the relationship between observed MPPGL values and source of liver tissue for preparation of microsomal fractions, the dataset was reduced to a set of samples encompassing an age range (3 years and 18 years of age (early childhood, middle childhood, and early adolescent age groups) that was common to each major source of tissues. This “trimmed” dataset included 92 samples from four tissue sources: UMBTB (n = 33), Minnesota (n = 31), Pittsburgh (n = 15), and Sekisui-Xenotech (n = 14); three samples within this age range obtained from Vitron were excluded from the analysis because of the small number of samples that fell within the specified age range. By univariate analysis, all four tissue sources were similar with respect to age and sex (∼40% female) but differed with respect to RNA quality; UMBTB samples had lower median (interquartile range) RQI values of 4.7 (3.3, 6.2) compared with 7.4 (6.0, 8.3) for Xenotech samples, 8.8 (8.2, 9.3) for Pittsburgh samples, and 8.9 (8.4, 9.3) for samples from the University of Minnesota source.
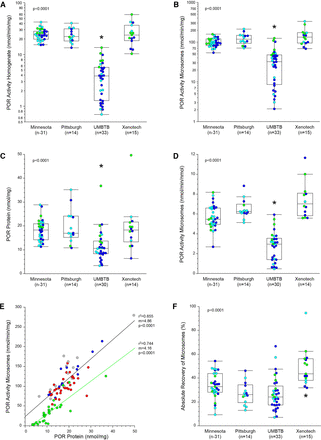
The potential tissue source effect on MPPGL was also assessed using POR activity and protein content as measures of microsomal quality. POR activity in liver homogenates and microsomes differed across the tissue sources as assessed by Welch’s ANOVA for unequal variances (P < 0.0001; POR activity in the UMBTB samples was lower than the other three sources; Figs. 2A and 2B). Proteomic analysis confirmed that POR protein was significantly lower in the UMBTB samples compared with the other three sources (P < 0.0001; Fig. 2C), and POR activity in microsomes expressed relative to absolute POR protein content was likewise significantly lower in the UMBTB samples (P < 0.0001; Fig. 2D). The relationship between microsomal POR activity and protein was similar for the UMBTB and non-UMBTB sources in terms of slope and coefficient of determination, but for a given amount of POR protein, activity was approximately 50% lower in the UMBTB samples (Fig. 2E). Finally, microsomal recovery also differed according to tissue source (Fig. 2F), with higher recovery from Sekisui-Xenotech samples (49.0 ± 15.8%; P < 0.001) compared with the other three sources, LTCDS-Minnesota (33.7 ± 10.6%), LTCDS-Pittsburgh (27.3 ± 9.4%) and UMBTB (27.0 ± 13.5%). Similar results were observed when all postnatal samples for the four sources were considered.
Effect of tissue source on POR activity and recovery of microsomes in the “trimmed” dataset. The dataset was reduced to a set of samples encompassing an age range, 3 years to 18 years of age, that was common to each major source of tissues. The effect of tissue source was assessed for POR activity per mg tissue homogenate protein (Panel A) and per mg microsomal protein (panel B), per nmol POR protein determined by quantitative proteomics (Panel C), and microsomal POR activity expressed relative to POR protein (Panel D). POR activity in microsomes correlated with microsomal POR protein content (Panel E), although activity was lower in UMBTB samples than non-UMBTB samples for a given amount of POR protein. The effect of tissue source on absolute recovery of microsomal protein is presented in Panel F. Data points in Panels A-D and F are color-coded by NIH age category: early childhood (green), middle childhood (turquoise) and early adolescence (blue). Data points in Panel E are color coded by tissue source as described in the legend to Fig. 1. Construction of box plots is as described in the legend to Fig. 1; the asterisk designates statistically significant difference by Tukey’s HSD as described in Methods.
As observed in the parent dataset, MPPGL values of the trimmed dataset were not normally distributed by visual inspection of the frequency histogram and Q-Q plots and the Shapiro-Wilk W test (P = 0.003). Therefore, the data were log-transformed and more closely approximated a normal distribution by visual inspection of the frequency histogram and Q-Q plots. Despite the lower POR activities in the UMBTB samples, comparison of logMPPGL values across the four sites revealed that values for the University of Minnesota samples were approximately 50% higher than the other groups by Tukey’s Honestly Significant Difference (P < 0.0001; Fig. 3).
Effect of tissue source on log-transformed MPPGL values in the trimmed dataset. Samples are color-coded by age category as described in the legend to Fig. 2. logMPPGL values were significantly greater in samples from the University of Minnesota compared with the other sources by Tukey’s HSD.
Relationship between MPPGL and Postnatal Age: Linear Regression Modeling.
After adjusting for source effects as described in Methods, linear models were fitted to the full data set (all sources), the data from all sources except UMBTB, and the UMBTB data alone (Table 2). The first two analyses were limited to ages ranging from 0 to 44 years to allow for full maturation to be achieved. There were only 16 samples in the 45-79 age range, all but one from a single source (Pittsburgh); these were excluded to avoid inordinate influence of data in the tail of the age distribution and over-reliance on data from a single source for this age range. The UMBTB analysis was limited to ages 0 to 25 by excluding two samples with outlying ages (35 and 47). In all models, age was centered at 9 years and expressed in decades. Source-adjusted log(MPPGL) was modeled as a function of sex, POR value (expressed as a z-score), age, and age squared. Respective medians for males and females were 1.34 and 1.36 for logMPPGL (0.1-SD difference, Wilcoxon 2-sample test P = 0.921), 14.6 and 19.6 nmol/min/mg protein for POR activity in homogenates, respectively, for males and females (0.36-SD difference, Wilcoxon P = 0.256), and 78.9 and 94.4 nmol/min/mg protein, respectively, for males and females for POR activity in microsomes (0.26-SD difference, Wilcoxon P = 0.177). Similarly, sex was not a significant factor in the initial model, and the linear model re-fit without sex, resulting in the results presented in Table 2. We explored cubic models, including natural spline models, for the full data set, but these models provided little or no additional explanatory value. Review of the results presented in Table 2 reveals that the 95% confidence intervals for some of the age variables (age and age2) include zero. The confidence intervals including zero are interpreted as zero (i.e., no change in logMPPGL with age) being a plausible value, given the data, for the true regression coefficient, along with every other value covered by the confidence interval.
Results of logMPPGL regression models
The prediction equations for each analysis (POR z-score assumed to be zero [i.e., average]) are as follows:
All sources Log(MPPGL) = 1.444 + 0.050*(age in decades – 0.9) - 0.010*(age in decades – 0.9)2
All sources except UMBTB (“non-UMBTB model”) logMPPGL = 1.515 + 0.001*(age in decades – 0.9) - 0.094*(age in decades – 0.9)2
UMBTB only (“UMBTB model”) logMPPGL = 1.069 + 0.070*(age.in.decades – 0.9) - 0.002* (age.in.decades – 0.9)2
The three models resulting from the regression analysis confirm that the UMBTB samples are driving the apparent age effects when all samples are considered together, as essentially no effect of age on logMPPGL is observed when the UMBTB samples are excluded from the analysis. Thus, the model provides an MPPGL value of 101.515 or 32.7 mg/g at the average age of 9 years for the age range of one month (the youngest sample in the non-UMBTB sample set) to 25 years, the oldest age included in the analysis; applying the non-UMBTB model to all samples from birth to 18 years results in a mean ± SD MPPGL value of 30.4 ± 1.7 mg/g (range 27.4 to 32.7 mg/g), similar to the value of 32.1 ± 4.3 mg/g (25.5 to 38.2 mg/g) that is obtained for the same samples using the equation of Barter et al. (Barter et al., 2007).
Relationship between MPPGL and Postnatal Age: Sigmoid Emax Model.
Application of a sigmoid Emax model to the same dataset used for the regression analysis failed to reach convergence on parameter estimates, consistent with minimal change in logMPPGL values from 1 month postnatal age to adulthood.
Discussion
Pediatric data describing the developmental trajectory of MPPGL, an important scaling factor for translating drug biotransformation activity in vitro into drug clearance values in vivo for PBPK modeling and simulation, are extremely limited. The goal of this investigation was to address this important knowledge deficit.
Compared with the availability of adult liver tissue, liver tissue from the pediatric age group (defined as birth to 18 years of age for this study and the age range of patients treated in children’s hospitals like our own) is much less common as every attempt will be made to find a suitable pediatric transplant recipient for any pediatric tissue that becomes available. Nevertheless, pediatric tissues for research are available from several academic and not-for-profit groups, such as UMBTB, LTCDS, the National Disease Research Interchange (NDRI; Bethesda, MD) and the Association of Human Tissue Users. Commercial sources of pediatric tissue or subcellular fractions include Xenotech (Kansas City, KS), Corning Gentest (Woburn, MA), Vitron (Tucson, AZ), Cellz Direct (Carlsbad, CA, USA), and PuraCyp (Carlsbad CA, USA), among others. Often, as in our case, tissues are acquired from multiple sources to accumulate sufficiently large numbers to generate robust and useful data, especially in neonates, toddlers and infants for whom extrapolation from adults becomes increasingly more tenuous.
This process, however, is accompanied by concerns related to the impact of variability between tissue sources and interpretation of the resulting data. For example, in the set of samples used for these studies, not all age groups are equally represented among all tissue sources. More specifically, 80% (16/20) of samples in the neonatal+infant age group (birth to 1 year of age) were obtained from a single source, UMBTB. Similarly, the age of the Pittsburgh samples (all adult) is much greater than the age of the Minnesota and UMB samples. Thus, the various tissue sources may differ with respect to the demographic characteristics of the population available through that source.
Further confounding the interpretation of the results is the fact that tissues are procured, maintained, and processed by substantially different procedures and protocols. At one extreme, liver tissue available from the University of Minnesota LTCDS site is snap frozen in liquid nitrogen within 60 minutes of explantation or ischemia start. This requirement is adhered to strictly and specimens outside of that window are disqualified for further use (Bartosz Grzywacz, MD, personal communication 4/6/2020). In this context, Barter et al. (Barter et al., 2008) found no difference in MPPGL values determined directly from a fresh tissue sample (21.9 ± 0.3 mg/g) compared with the value determined after thawing a matching snap-frozen sample (23.5 ± 1.2 mg/g). The University of Maryland Brain and Tissue Bank for Developmental Disorders was founded in 1991 in response to the need for increased research on developmental disorders impacting children. Now part of the NIH NeuroBioBank network, the UMBTB continues to receive tissue donations from families; after the consent process has been completed, tissues are harvested by a medical examiner or forensic pathologist and rinsed with water before freezing and transfer to the Biobank, essentially autopsy samples (https://www.medschool.umaryland.edu/btbank/Medical-Examiners-and-Pathologists/ Consent-and-Tissue-Recovery-Process/). A third procedure used by commercial entities, such as Sekisui Xenotech, involves tissues obtained through partnerships with non-profit organ procurement organizations managed by the regional Organ Procurement and Transplantation Networks that are regulated by the United Network for Organ Sharing (UNOS; Richmond, VA) and ultimately deemed unsuitable for transplant. After harvest, tissues are perfused with a preservation buffer and kept on wet ice until unpacked by the recipient organization and flash frozen. Continual improvement in the preservation solutions is a science of its own, with the goal of minimizing ischemia damage and increasing transplant success (Petrenko et al., 2019).
Although tissues available through UMBTB had slightly shorter post-mortem intervals (12.1±3.1 hour) compared with organizations acquiring livers preserved in some manner between explantation and acquisition (Sekisui-XenoTech 15.1±5.9 hour; University of Pittsburgh 15.6±9.2 hour), differences in RNA quality were apparent, with the UMBTB samples having lower RQI values than the other three primary sources of tissues. Similar differences were observed for POR activity, selected as the microsomal marker activity as considerably less sample is required compared with the amount required for carbon monoxide binding spectra, an important consideration given the limited amount of tissue (∼1 g or less) available for some samples. POR activity correlated with POR protein abundance determined by quantitative proteomic determination and POR activity, (r2=0.679, P < 0.0001), but lower activity and protein abundance were observed in the UMBTB samples relative to the other sources, providing confidence in POR activity as a measure of tissue quality and marker of microsomal content and recovery.
To the extent that POR activity in liver homogenates (and subsequently microsomes) can be used as a measure of tissue quality, UMBTB/autopsy livers in the “trimmed” dataset used for assessing source effects had considerably lower POR activity (and microsomal POR protein abundance) than the other sources. However, as calculation of MPPGL essentially is based on the ratio of PORHom and PORMic (Eq. 3), MPPGL values for the UMBTB samples tended to be lower and more variable than the Sekisui-Xenotech and Pittsburgh samples such that it was difficult to detect a difference among the three groups (Fig. 3). Taking the considerations above into account, we conclude that liver tissue samples acquired through UMBTB are different from the LTCDS sites at the Universities of Minnesota and Pittsburgh and Sekisui-Xenotech, and some of the differences in POR activities may be due to tissue quality. However, we note that as UMBTB was originally developed to serve as a resource for research in developmental disorders primarily affecting children, a state of chronic illness preceding death may also contribute to the observed differences in POR activities in UMBTB samples relative to other groups. On the other hand, estimation of MPPGL in UMBTB tissues based on the ratio of POR values implies that MPPGL values in these samples are within the range of values derived from the other major sources. Furthermore, among the pediatric samples, UMBTB samples are over-represented in the younger age groups (infant and toddler stages, in particular).
Given the effect of tissue source on POR activity and thus, MPPGL values, a value of 30.4±1.7 is representative of an age range of one month to early adulthood resulting from this analysis (geometric mean of 28.0±1.4 mg/g for the non-UMBTB trimmed data set) and represents the best estimate of MPPGL for the pediatric age range. We acknowledge that there may be different approaches to investigate the effect of age on MPPGL (or lack thereof), and therefore, the MPPGL value for each sample included in this analysis is provided in Supplemental Table 2. Nevertheless, the value of 30.4±1.7 mg/g that we report is similar to the value of 32–33 mg/g reported by Barter et al. (Barter et al., 2007; Barter et al., 2008), and somewhat higher than the value of 18.7±2.8 mg/g derived from four pediatric liver samples obtained from infants/young children with biliary atresia as reported by De Bock et al. (De Bock et al., 2014). This latter value is striking in its similarity to the geometric mean value of 18.6±1.6 mg/g for the UMBTB samples (and higher than a value of 11.7 mg/g at age 9 years using the regression equation for UMBTB samples) in our study, possibly reflecting a disease-related effect, although not necessarily directly affecting the liver of patients suffering from chronic disease. Beyond estimates of average values of MPPGL for use in modeling and simulation, an understanding of the inter-individual variability in MPPGL values at a given age or developmental stage is perhaps equally important, generally varying approximately 4- to 6-fold in the postnatal age groups included in the study.
In summary, this report addresses an important knowledge deficit regarding the ontogeny of a critical scaling factor for extrapolating in vivo drug clearance in pediatric age groups from in vitro drug biotransformation data. The analysis also identified tissue source and quality as potential factors influencing interpretation of the data. During preparation of this manuscript, a paper was published by Doerksen et al. in which several important considerations related to MPPGL and the related factor cytosolic protein per gram liver are critically reviewed (Doerksen et al., 2021). These authors also emphasize the importance of tissue source, and identify methodological considerations, such as buffers and tissue processing protocols, choice of protein assay, and marker for calculating recovery (e.g., CYP content, POR activity) as additional factors of importance. In addition to the source-dependent effect on POR reported in this paper, we have observed similar effects on CYP3A4 activity (and other CYPs; unpublished data) and naltrexol formation (attributed to aldo-keto reductase family 1, member C4) in cytosols prepared from the same liver samples (Stancil et al., in press). Given the limited number of quality pediatric tissues available for research, standardization of efforts to generate high-quality pediatric data from multiple tissue sources will lead to refinement of developmental trajectories beyond those currently available (for example, UMBTB samples reported in Stevens et al., 2003 and Koukouritaki et al., 2004) and ultimately, improved modeling and simulation for pediatric applications.
Authorship Contributions
Participated in research design: Leeder, Dinh, Pearce.
Conducted experiments: Gaedigk, Prasad, Pearce.
Contributed new reagents or analytic tools: Leeder, Prasad, Pearce.
Performed data analysis: Leeder, Staggs.
Wrote or contributed to the writing of the manuscript: Leeder, Dinh, Gaedigk, Staggs, Prasad, Pearce.
Footnotes
- Received July 22, 2021.
- Accepted October 20, 2021.
↵1 Current affiliation: Certara, Princeton, New Jersey.
Tissues used in this study were acquired from publicly supported tissue repositories, including the National Institutes of Health (NIH)-funded University of Maryland Brain and Tissue Bank for Developmental Disorders (funded by NIH contract HHSN275200900011C, Ref. No. #N01-HD-9-0011) and The Liver Tissue Cell Distribution System (funded by NIH contract #N01-DK-7-0004/HHSN267200700004C). The work was supported by grants P50 HD090258 (J.S.L.) and R01-HD081299 (B.P.) from the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
No author has an actual or perceived conflict of interest with the contents of this article.
↵
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
Abbreviations
- LTCDS
- Liver Tissue Cell Distribution System
- MPPGL
- microsomal protein per gram liver
- PBPK
- physiology-based pharmacokinetic
- PK
- pharmacokinetic
- POR
- cytochrome P450 oxidoreductase
- UMBTB
- University of Maryland Brain and Tissue Bank for Developmental Disorders
- AHTU
- Association of Human Tissue Users
- AKR1C4
- aldo-keto reductase family 1, member C4
- CPPGL
- cytosolic protein per gram liver
- HLM
- human liver microsomes
- LC/MS/MS
- liquid chromatography with tandem mass spectroscopy
- Copyright © 2021 by The American Society for Pharmacology and Experimental Therapeutics