Abstract
The levels of drug-metabolizing enzymes (DMEs) and transporter proteins in the human intestine are pertinent to determine oral drug bioavailability. Despite the paucity of reports on such measurements, it is well recognized that these values are essential for translating in vitro data on drug metabolism and transport to predict drug disposition in gut wall. In the current study, clinically relevant DMEs [cytochrome P450 (P450) and uridine 5′-diphospho-glucuronosyltransferase (UGT)] and drug transporters were quantified in total mucosal protein preparations from the human jejunum (n = 4) and ileum (n = 12) using quantification concatemer–based targeted proteomics. In contrast to previous reports, UGT2B15 and organic anion-transporting polypeptide 1 (OATP1A2) were quantifiable in all our samples. Overall, no significant disparities in protein expression were observed between jejunum and ileum. Relative mRNA expression for drug transporters did not correlate with the abundance of their cognate protein, except for P-glycoprotein 1 (P-gp) and organic solute transporter subunit alpha (OST-α), highlighting the limitations of RNA as a surrogate for protein expression in dynamic tissues with high turnover. Intercorrelations were found within P450 [2C9-2C19 (P = 0.002, R2 = 0.63), 2C9–2J2 (P = 0.004, R2 = 0.40), 2D6-2J2 (P = 0.002, R2 = 0.50)] and UGT [1A1-2B7 (P = 0.02, R2 = 0.87)] family of enzymes. There were also correlations between P-gp and several other proteins [OST-α (P < 0.0001, R2 = 0.77), UGT1A6 (P = 0.009, R2 = 0.38), and CYP3A4 (P = 0.007, R2 = 0.30)]. Incorporating such correlations into building virtual populations is crucial for obtaining plausible characteristics of simulated individuals.
SIGNIFICANCE STATEMENT A number of drug transporters were quantified for the first time in this study. Several intercorrelations of protein abundance were reported. mRNA expression levels proved to be a poor reflection of differences between individuals regarding the level of protein expression in gut. The reported abundance of drug-metabolizing enzymes and transporters and their intercorrelations will contribute to better predictions of oral drug bioavailability and drug-drug interactions by linking in vitro observations to potential outcomes through physiologically based pharmacokinetic models.
Introduction
Physiologically based pharmacokinetic (PBPK) models are increasingly used to predict drug disposition, efficacy, and toxicity across different patient populations. However, to achieve reliable information from PBPK models, they need to be populated with accurate experimental data, which is essential to the translation of in vitro data to anticipate in vivo observations (Rostami-Hodjegan, 2012). Information regarding the abundance of drug-metabolizing enzymes (DMEs) and drug transporters in relevant human organs is crucial to building and refining PBPK models, consequently aiding the assessment of the performance of existing and new drugs in different patient populations and across different pathologies (Rostami-Hodjegan, 2012; Jamei, 2016).
The main organ responsible for drug metabolism in the human body is the liver; however, orally administered drugs need to cross the gastrointestinal tract, mainly through absorption in the small intestine, into the portal vein before arriving at the liver. The absorption of drugs from the small intestine is not only influenced by different families of DMEs, such as cytochrome P450 (P450) and uridine 5′-diphosphate-glucuronosyltransferase (UGT), but also by drug transporters from the ATP-binding cassette (ABC) and solute carrier (SLC) families. However, the distribution of these enzymes and transporters may vary across different regions in the small intestine because these regions have distinct physiologic roles. The physiologically variable and dynamic environment in the gut lumen together with the high turnover of enterocytes (Darwich et al., 2014, 2019) may play a role in the high interindividual variation observed in the abundance of these proteins (Paine et al., 2006). Thus, the prediction of the absorption and gut wall clearance of orally administered drugs is considereddifficult (Sjögren et al., 2014). Therefore, to make meaningful progress toward achieving accurate PBPK predictions of absorption and gut wall clearance of orally administered drugs from the small intestine, it is vital that the abundance of DMEs and drug transporters and their variations are quantified in the small intestine. To capture the variability in a population, quantification of these protein abundances should be achieved in a sufficient number of individuals across different patient populations (Drozdzik et al., 2018; Harwood et al., 2019). The invasive nature of procedures and limited access to dedicated intestinal tissue banks has so far precluded such a large-scale study. Despite this challenge, published data do exist for DMEs and transporters in a relatively small number of samples (Gröer et al., 2013; Sato et al., 2014; Akazawa et al., 2018; Harwood et al., 2019). Regional variations in the abundance of some DMEs and drug transporters have also been reported in relatively few studies, whereas no variations were observed in other studies (Drozdzik et al., 2014, 2018). Challenges associated with obtaining intestinal tissue samples increase the value of studies reporting the actual protein abundances as opposed to mRNA levels. There are a number of reports on mRNA expression data of genes encoding DMEs and drug transporters in the human small intestine (Thörn et al., 2005; Clermont et al., 2019; Fritz et al., 2019). However, caution should be exercised in using these values because the relationship between mRNA and protein abundance appears inconsistent (Berggren et al., 2007; Hayeshi et al., 2008), suggesting that mRNA data in a high-turnover tissue such as intestine may have limited utility for PBPK modeling. In contrast, evidence of a good correlation between protein abundance and protein activity has recently been reported for DMEs and transporters (Achour et al., 2017; Couto et al., 2019; El-Khateeb et al., 2019) in other organs, such as liver. This suggests that the availability of abundance data for these proteins will be key to developing robust PBPK models for oral drug absorption.
Using quantification concatemer (QconCAT)-based proteomics and a well established proteomic workflow, we aimed to quantify the protein abundance of key P450 (2C9, 2C19, 2D6, 2J2, and 3A4) and UGT (1A1, 1A3, 1A6, 2B7, and 2B15) enzymes in two regions of the human small intestine: the jejunum and ileum. Simultaneously, several transporters from the SLC and ABC protein families [(P-gp, breast cancer resistance protein (BCRP),multidrug resistance-associated protein 2 (MRP2), organic solute transporter subunit alpha (OST-α), organic solute transporter subunit beta (OST-β),organic anion transporter polypeptide-related protein 2 (OATP2B1). ) were also quantified. In addition, the mRNA expression levels of relevant transporters were measured in the same samples, enabling the assessment of correlations between mRNA expression and protein abundance of transporters. Because of the relatively large number of samples in comparison with existing proteomics studies of DMEs and drug transporters in human intestine, the intercorrelation between the protein abundance of enzymes and transporters was also investigated. The impact of factors such as age and sex on the expression of drug-metabolizing enzymes and transporters was also evaluated.
Materials and Methods
Chemicals.
Unless otherwise indicated, all chemicals were supplied by Sigma-Aldrich (Poole, UK) with the highest purity available. All solvents were high-performance liquid chromatography (HPLC) grade and supplied by ThermoFisher Scientific (Paisley, UK). Unlabeled peptide standard EGVNDNEEGFFSAR ([Glu1]-Fibrinopeptide B) and its modified analog GVNDNEEGFFSAR, each with 95% purity, were purchased from Severn Biotech Ltd. (Worcestershire, UK). Lysyl endopeptidase, mass spectrometry grade, was purchased from Wako (Osaka, Japan). Sequencing-grade modified trypsin was supplied by Promega (Southampton, UK). Complete Mini, EDTA-free protease inhibitor cocktail tablets were supplied by Roche (Mannheim, Germany). The DNA constructs for the QconCATs were produced by PolyQuant GmbH (http://www.polyquant.com/) (Rogensburg, Germany) using the expression vector pET21a encoding ampicillin and kanamycin resistance.
Human Intestinal Samples.
Human intestinal tissues (16 donors) were obtained after receiving informed consent from patients undergoing intestinal surgery at Salford Royal NHS Foundation Trust, Salford, UK. Prior ethics committee approval had been granted by the North West Research Ethics Committee, UK (06/1410/126), and all procedures were carried out in accordance with the Declaration of Helsinki guidelines. Samples were removed from the healthy margins of resected specimens at a minimum of 5 cm from the diseased mucosa and were macroscopically normal. Following removal of the intestinal sample, mucosal tissue was rapidly separated from muscle and serosal layers by blunt dissection. Samples of mucosa (∼1 cm2) were then snap frozen in liquid N2 and stored at −80°C for subsequent analysis of protein abundance and mRNA expression. Donor demographic (ethnicity, age, and gender) and clinical (medical history and medications) information are provided in Supplemental Table 1. An overview of the experimental workflow is shown in Supplemental Fig. 1.
Tissue Preparation and Isolation of Total Human Mucosal Protein.
Snap-frozen mucosal tissue samples were ground to powder using a cooled mortar and pestle. This was resuspended in radioimmunoprecipitation assay (RIPA) lysis buffer containing 150 mM NaCl, 1% (v/v) Triton X-100, 0.5% (w/v) sodium deoxycholate, 0.1% (w/v) SDS, and 50 mM Tris base pH 8.0. The buffer also contained protease inhibitors (0.5 mM phenylmethanesulfonylfluoride and 50 µl ml−1 protease inhibitor cocktail). Following incubation and mixing at 4°C for 30 minutes, the extract was centrifuged at 14,000g for 5 minutes at 4°C. The supernatant was removed and stored in aliquots at −80°C. RIPA buffer was used for protein extraction because it has been shown to be a stringent lysis buffer capable of solubilizing cellular membranes to enable quantification of proteins localized at the endoplasmic reticulum and plasma membrane (Padilla-Benavides et al., 2010; Feng et al., 2015). The use of RIPA buffer followed by centrifugation is not a membrane protein enrichment technique. Therefore, the protein samples extracted using this strategy are representative of total cellular protein.
QconCAT Design and Expression.
Two different QconCATs previously designed to quantify human hepatic transporters (Concatemer of Standard Peptides from Human Hepatic Transporters; TransCAT) and human hepatic metabolizing enzymes (Concatemer of Standard Peptides from Human Drug Metabolizing Enzymes; MetCAT) were used to quantify the same transporters and DMEs from human intestinal tissues (Russell et al., 2013; Harwood et al., 2015). From the MetCAT construct, two unique peptides belonging to each of five CYP450s (CYP2C9, CYP2C19, CYP2D6, CYP2J2, and CYP3A4) and five UGTs (UGT1A1, UGT1A3, UGT1A6, UGT2B7, and UGT2B15) were selected for quantification. To enable accurate quantification of the MetCAT, [Glu1]-Fibrinopeptide B analog (GVNNEEGFFSAR) omitting the N-terminal glutamate residue was incorporated in the sequence. The TransCAT construct was designed to incorporate two unique peptides, each belonging to specific transporter proteins of the ABC family, i.e., P-gp, BCRP, and MRP2 and the SLC superfamily OST-α, OST-β, OATP1A2, and OATP2B1. In addition, two plasma membrane marker proteins, sodium/potassium-transporting ATPase subunit alpha-1 (Na+/K+-ATPase) and human peptide transporter 1 cadherin-17 (CDH-17)], were included in this construct. To enable quantification of the TransCAT, F, [Glu1]-Fibrinopeptide B B peptide sequence (EGVNDNEEGFFSAR) was also incorporated in the TransCAT sequence. The peptides selected for quantification are shown in Supplemental Tables 2 and 3. Expression of heavy isotope–labeled MetCAT and TransCAT in Escherichia coli was carried out as previously described (Russell et al., 2013).
Protein Content Quantification.
Protein content of the human intestinal extracts was estimated using Bradford protein assay according to the manufacturer’s instructions. This involved Bio-Rad based on the Coomassie Brilliant Blue G-250 dye (ThermoFisher Scientific, Hemel Hempstead, UK). The analysis was made in triplicate according to the manufacturer’s protocol using bovine serum albumin as a standard. To minimize the effect of RIPA buffer components on the determination of protein content in the total mucosal protein extracts, mucosal intestine samples were diluted 100 times in HPLC water before the assay was performed.
RNA Extraction and qRT-PCR.
Snap-frozen tissue samples were ground to a powder, and total RNA was extracted by resuspending in Tri-Reagent (Thermo Fisher Scientific) and processing using standard procedures. Following determination of the concentration and purity of isolated RNA by A260/A280 spectrophotometry, cDNA was prepared from 3 µg of total RNA in a total volume of 20 µl using the Roche Transcriptor First Strand cDNA Synthesis kit (Roche, Burgess Hill, UK).
Relative quantification of gene expression was undertaken for seven transporters (ABCB1, ABCG2, ABCC2, SLCO2B1, SLC51A, SLC51B, and CDH-17) by real-time polymerase chain reaction using the Roche Universal Probe Library (UPL) system (Roche). Using the UPL Genefinder software, gene-specific intron-spanning primers and appropriate fluorescent hydrolysis probes were designed for each transporter. Assays were performed using the Roche Lightcycler 480 platform in a total volume of 20 µl with 200 nM forward and reverse primers, 100 nM of the UPL hydrolysis probe, and ∼0.3 µg cDNA. The comparative threshold cycle method was used to determine mRNA expression relative to reference genes glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and villin 1. Sequences of the polymerase chain reaction primers are supplied in Supplemental Table 4.
Total Mucosal Protein Digestion.
To enable quantification by mass spectrometry, 20 µg of each total mucosal protein fraction was spiked with a known amount of isotope-labeled MetCAT and TransCAT. To each fraction containing the MetCAT and TransCAT standards, sodium deoxycholate was added to a final concentration of 5% (w/v). The mixture was thoroughly mixed and incubated at room temperature for 10 minutes.
For protein digestion, the filter-aided sample preparation method (Wiśniewski et al.,2009) was adopted as previously described (Al Feteisi et al., 2018; Al-Majdoub et al., 2019; Couto et al., 2019). Briefly, the detergent-solubilized proteins from the human total mucosal protein samples were reduced using 100 mM 1,4-dithiothreitol, followed by alkylation with 50 mM iodoacetamide. After alkylation, deoxycholate removal was performed by buffer exchange using successive washes with 8 M urea in 100 mM Tris-HCl (pH 8.5). To reduce urea concentration, three additional washes were performed using 1 M urea in 50 mM ammonium bicarbonate (pH 8.5). Protein digestion was achieved by sequential treatment with endopeptidase Lys-C and trypsin (Al-Majdoub et al., 2014). Firstly, Lys-C was added at an enzyme:protein ratio of 1:50, which was followed by incubation at 37°C for 4 hours. After Lys-C, trypsin was added at a ratio 1:25 enzyme:protein, which was followed by overnight incubation at 37°C. Peptides were recovered by centrifugation (14,000g, 20 minutes), followed by elution using 0.5 M sodium chloride. Unlabeled peptide standard EGVNDNEEGFFSAR ([Glu1]-Fibrinopeptide B) and its modified analog GVNDNEEGFFSAR were added and desalted using a C18 column (Thermo). The peptide mixtures were lyophilized using a vacuum concentrator and stored at −20°C until mass spectrometric analysis.
Liquid Chromatography and Mass Spectrometry.
Proteins were quantified by mass spectrometry targeted proteomics based on a published protocol (Harwood et al., 2015). For all analyte peptides and their isotope-labeled internal QconCAT peptides, three mass transitions were selected (see Supplemental Tables 2 and 3). Multiple reaction monitoring (MRM) acquisition was applied for optimization of transitions using a pooled sample. Each individual sample was injected in duplicate into the liquid chromatography system.
Briefly, dried peptide samples were resuspended in loading buffer [HPLC-grade water containing 5% (v/v) HPLC-grade acetonitrile and 0.1% (v/v) formic acid] to reach a concentration of 1 µg µl−1, and 1.0 µl of each sample was loaded on an UltiMate 3000 rapid separation liquid chromatography (Dionex, Surrey, UK) coupled to an on-line QTRAP 6500 mass spectrometer (SCIEX, Warrington, UK). Peptides were reverse-phase separated on a PepMap rapid separation liquid chromatography C18 column (2-µm particles, 100 Å, 75-µm inner diameter, 50-cm length; Thermo Scientific) preceded by a C18 PepMap100 µ-Precolumn (5 µm, 100 Å, 5-µm inner diameter, 5-mm length; ThermoFisher Scientific). A multi-step gradient was used from 4% to 40% buffer B [80% (v/v) acetonitrile with 0.1% (v/v) formic acid] for 60 minutes at a flow rate of 300 nl min−1. The composition of buffer A was HPLC-grade water containing 0.1% (v/v) formic acid.
Assessment of P450, UGT, and Transporter Abundance.
Protein/peptide quantification was performed using Skyline v4.0 (https://skyline.ms/project/home/software/Skyline/begin.view) (MacLean et al., 2010). For each peptide of interest, MRM transitions were optimized using a pooled sample in a single analytical run. Two unique peptides and three transitions were used for the quantification of all CYP450 and UGT enzymes, except CYP3A4, UGT1A6, and UGT2B7, which were quantified using a single peptide. From the UGT enzymes, isomeric sequences, such as VSVWLLR and ADVWLIR, which are unique to UGT1A6 and UGT2B7, respectively, were not considered because of the overlapping retention time and interference of signal intensities. All transporters were quantified using a single peptide and three transitions, except OATP2B1, which was quantified using a single peptide and two transitions only. Single peptides were used for quantification because the elution profile of the second peptides (in the heavy and light versions) was not of adequate quality to be considered for quantification. The peptide sequences used for quantification are provided in Supplemental Tables 2 and 3. Representative examples of MRM coelution profiles for all transporters are shown in Supplemental Figs. 2 and 3. For each selected targeted peptide, concentration was calculated based on the mean of at least two mass transitions per peptide.
Statistical Analysis.
All statistical analyses of the data were performed using Microsoft Excel 2010 and GraphPad Prism v7.03 (La Jolla, CA). Nonparametric statistics were used because a considerable proportion of the data set did not follow normal distribution. The normality of data distribution was assessed using three tests: D’Agostino-Pearson, Shapiro-Wilk, and Kolmogorov-Smirnov normality tests. All protein expression data were presented as means ± S.D., CV, and minimum and maximum. The Spearman rank-order correlation (Rs) test with t-distribution of the P values was used to assess mRNA-protein correlation and intercorrelation between protein abundance levels. The level of scatter of data was evaluated by linear regression (R2). Correlations were considered strong when the values correlated well (Rs > 0.60), with statistical significance P value less than 0.05, and demonstrated limited scatter (R2 > 0.30). Rs values between 0.5 and 0.6 with a P value less than 0.05 were considered to represent a moderate correlation. The relationship between age and expression level was also assessed using these correlation tests. Sample differences were evaluated using the nonparametric Mann-Whitney U test and Kolmogorov-Smirnov cumulative distribution test. The relationship between protein abundance levels and the expression covariates, sex and disease, was assessed using Mann-Whitney U test and Kruskal-Wallis one-way ANOVA.
Results
In this study, the MetCAT standard was used to measure the abundance of clinically relevant DMEs from the CYP450 family (CYP2C9, CYP2C19, CYP2D6, CYP2J2, and CYP3A4) and UGT family (UGT1A1, UGT1A3, UGT1A6, UGT2B7, and UGT2B15) across 16 human intestine samples, four of which were from the jejunum and 12 of which were from the ileum. In the same experiment, the TransCAT was used to measure protein abundance of a membrane marker (Na+/K+-ATPase), relevant drug transporters from the ABC (P-gp, BCRP, and MRP2) and SLC (OST-α, OST-β, OATP1A2, and OATP2B1) families, and cadherin-17, a cell adhesion molecule that also functions as a proton-dependent oligopeptide transporter in the human intestine (Dantzig et al., 1994). In addition to measurement of protein abundance, the relative mRNA expression levels of P-gp (ABCB1), MRP2 (ABCC2), BCRP (ABCG2), OST-α (SLC51A), OST-β (SLC51B), OATP2B1 (SLCO2B1), and CDH-17 were measured. The experimental workflow is shown in Supplemental Fig. 1.
Abundance of P450 and UGT Enzymes in Human Intestinal Samples.
Table 1 and Fig. 1 summarize the abundance of P450 and UGT enzymes in four jejunum and 12 ileum samples. All targeted P450 enzymes were expressed in both jejunum and ileum following the rank order CYP3A4 > CYP2D6 > CYP2C9 > CYP2C19 > CYP2J2, with CYP3A4 being the most abundant in both the jejunum and ileum and CYP2D6 being the second most abundant (yet one order of magnitude lower and relatively close to the abundances of CYP2C9, CYP2C19, and CYP2J2). A marginally higher expression of CYP2C9, CYP2D6, CYP2J2, and CYP3A4 was found in the jejunum compared with the ileum, whereas for CYP2C19, this relationship is reversed. With the exception of CYP2J2, the abundance of P450 enzymes is more heterogeneous in the ileal samples compared with the jejunum (Table 1). With the exception of CYP3A4 (Mann-Whitney U test, P = 0.03), no statistically significant differences were observed between quantified P450 abundances in jejunum and ileum.
Expression levels of enzymes (P450 and UGT) with known involvement in drug metabolism in human intestine
Protein expression is represented by the mean, the S.D. of the mean, the %CV, and the range (min-max). Protein abundance is reported in picomoles per milligram of total mucosal protein.
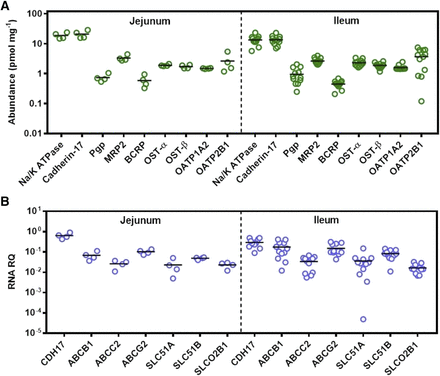
Abundance of drug-metabolizing enzymes in the human intestine. In (A and B), scatter plots represent the abundance of drug-metabolizing P450 and UGT enzymes, respectively, in 16 human intestinal samples from jejunum (n = 4) and ileum (n = 12). Protein abundances are expressed as picomoles per milligram of total mucosal protein from intestinal tissue. Bars in black represent the mean.
The quantified UGT abundance levels followed the rank order UGT1A1 > UGT2B7 > UGT1A3 > UGT1A6 > UGT2B15 in both jejunum and ileum. UGT1A1 was the most abundant UGT protein, followed by UGT2B7 and UGT1A3 (Fig. 1B; Table 1); UGT2B15 was the least abundant in both intestinal regions. To the best of our knowledge, this is the first report of UGT2B15 abundance quantification in the human small intestine (Miyauchi et al., 2016). In general, the average protein abundance of all UGTs tended to show a trend toward higher expression in the jejunum than in ileum; however, no statistically significant differences were observed.
Protein Abundance and mRNA Transcript Abundance of ABC and SLC Transporters in Human Intestinal Samples.
With respect to measured abundances of transporters (Fig. 2A; Table 2), ABC transporters followed the rank order MRP2 > P-gp ≥ BCRP in both jejunum and ileum. In the SLC family of transporters, OATP2B1 was the most abundant protein, followed by OST-α, OST-β, and OATP1A2. In previous targeted proteomic studies, quantifying OATP1A2 above limits of quantification was challenging in intestinal samples (Gröer et al., 2013; Drozdzik et al., 2014; Nakamura et al., 2016). A recent meta-analysis assessing relative and absolute regional expression of transporters in the intestine highlighted the challenges for assigning OATP1A2 expression directly into PBPK models in which a mechanistic intestinal model is described (Harwood et al., 2019). In this study, however, the abundance of OATP1A2 in all 16 samples was found to be above the limit of quantification at relatively similar levels to OST-α and OST-β. No statistically significant differences in protein abundance between jejunum and ileum were observed for any of the ABC and SLC transporters measured. The only proteins showing significant differences between the two regions were cadherin-17 and Na+/K+-ATPase. The abundance of both proteins was higher in jejunum, with the difference considered significant for cadherin-17 (Mann-Whitney U test, P = 0.03) and moderately significant for Na+/K+-ATPase (Mann-Whitney U test, P = 0.058).
Quantification of protein abundance and mRNA expression of drug transporters in the human intestine. In (A), scatter plots represent the protein abundance of relevant drug transporters from the ABC and SLC families, as well as the plasma membrane markers Na+/K+-ATPase and cadherin-17, in 16 human intestinal tissue samples from jejunum and ileum. Protein abundances are expressed as picomoles per milligram of total mucosal protein from intestinal tissue. In (B), scatter plots represent relative mRNA expression of selected drug transporters and the plasma membrane marker CDH-17 normalized to mRNA transcript levels of GAPDH in human intestinal tissue. RNA data were not collected for the gene ATP1A1 (corresponding to the protein Na+/K+-ATPase) and gene SLCO1A2 (protein OATP1A2). Proteins are denoted with their most common names, and transcripts are denoted by their corresponding gene names. The protein cadherin-17 corresponds to CDH-17, P-gp corresponds to ABCB1, MRP2 corresponds to ABCC2, BCRP corresponds to ABCG2, OST-α and OST-β correspond to SLC51A and SLC51B, respectively, and OATP2B1 corresponds to SLCO2B1. Bars in black represent the mean. RQ stands for relative quantification.
Expression levels of one plasma membrane marker (Na+/K+-ATPase), one cell adhesion protein (cadherin-17), and drug transporters with known involvement in drug clearance in human intestine
Protein expression is represented by the mean, the S.D. of the mean, the %CV, and the range (min-max). Protein abundance is reported in picomoles per milligram of total mucosal protein.
mRNA expression relative to the reference gene GAPDH was determined for all transporters with the exception of SLCO1A2 (OATP1A2) and ATP1A1 (Na+/K+-ATPase). Figure 2B summarizes transporter mRNA expression and protein abundance levels of the cell-cell adhesion/peptide transporter CDH-17, the ABC transporters [ABCB1 (P-gp), ABCC2 (MRP2), and ABCG2 (BCRP)], and solute carrier transporters [SLC51A (OST-α), SLC51B (OST-β), and SLCO2B1 (OATP2B1)] in jejunum and ileum for the 16 human intestinal samples. CDH-17 exhibited the highest values for protein abundance and mRNA expression in both jejunum and ileum. However, the overall rank order of mRNA expression is markedly different to that of protein abundance. No statistically significant differences were observed between mRNA expression levels in the jejunum and ileum.
Correlation between Protein Abundance of P450 and UGTs.
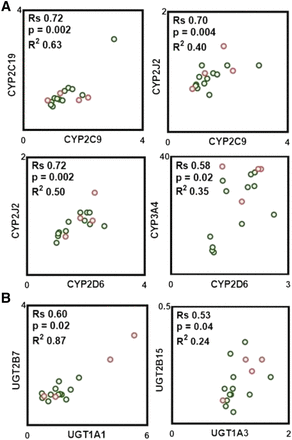
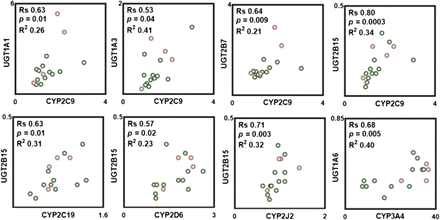
Pairwise correlation in protein abundance within P450, UGTs, and between P450 and UGTs was tested with the nonparametric Spearman rank test. As shown in Fig. 3, strong, significant, and positive correlations were found only between select pairs of P450, such as CYP2C9-CYP2C19, CYP2C9-CYP2J2, and CYP2D6-CYP2J2, whereas a moderate correlation was found between abundances of CYP2D6-CYP3A4. Analysis of correlations between UGT abundances indicates that a strong, significant, and positive correlation was only observed between UGT1A1-UGT2B7, whereas a moderate correlation was found between UGT1A3-UGT2B15. In addition, strong, significant, and positive correlations were found between protein abundances of certain P450 and UGT enzymes (Fig. 4). The abundance of CYP2C9 correlates well with the abundances of UGT1A1, UGT1A3, UGT2B7, and UGT2B15. CYP2C19 and CYP2J2 abundances also correlated well with UGT2B15. A moderate correlation was found between the abundance of CYP2D6 and UGT2B15. In general, CYP3A4 does not correlate with the abundance of any of quantified UGTs with the exception of UGT1A6, in which a strong correlation was found (Fig. 4).
Observed significant correlations in protein abundances between P450 (A) and between UGTs (B). Correlation analysis was performed using Spearman rank-order correlation (Rs) test with t-distribution of the P value. Linear regression analysis was carried out to assess the linearity of relationships and scatter of the data (R2). Correlations were deemed strong when the Rs values were greater than 0.60, P values were less than 0.05, and the data points demonstrated limited scatter (R2 > 0.30). Rs values between 0.5 and 0.6 with a P value less than 0.05 were considered to represent a moderate correlation. Clear circles with a green border correspond to protein abundance in the ileum, and clear circles with a red border correspond to protein abundance in the jejunum.
Observed correlations in protein abundances between P450 and UGT enzymes. Correlation analysis was performed using Spearman rank-order correlation (Rs) test with t-distribution of the P value. Linear regression analysis was carried out to assess the linearity of relationships and scatter of the data (R2). Correlations were deemed strong when the Rs values were greater than 0.60, P values were less than 0.05, and the data points demonstrated limited scatter (R2 > 0.30). Rs values between 0.5 and 0.6 with a P value less than 0.05 were considered to represent moderate correlation. Clear circles with a green border correspond to protein abundance in the ileum, and clear circles with a red border correspond to protein abundance in the jejunum.
Correlation between Protein Abundance of Transporters, P450, and UGTs.
Pairwise correlation in abundance within transporters and between transporters and DMEs was tested with the nonparametric Spearman rank test. A positive, strong and significant correlation was only observed between P-gp-OST-α (P < 0.0001, R2 = 0.77). The correlation between the abundance of transporters and DMEs was predominantly between P-gp-UGT1A6 (P = 0.009, R2 = 0.38) and MRP2-UGT1A1 (P = 0.01, R2 = 0.30). A moderate correlation between transporters and P450 was only observed for OATP1A2-CYP2J2 (P = 0.01, R2 = 0.20). A strong correlation was observed between P-gp and CYP3A4 (P = 0.007, R2 = 0.30, n = 15).
Correlation between Protein and mRNA of the Quantified Transporters.
Pairwise correlation between mRNA expression and protein abundance within transporters was tested with the nonparametric Spearman rank test. Significant correlations were observed for only two transporters (ABCB1/P-gp and SLC51A/OST-α), although the latter showed a high level of scattering between data points (Fig. 5A). Similar data were obtained using a second reference gene (villin 1) to determine relative mRNA expression (Fig. 5B).
Observed correlations between transporter protein abundance and relative mRNA expression levels. In (A), the x-axis represents mRNA expression levels of transporters normalized to mRNA expression levels of GAPDH, and the y-axis represents protein abundance. In (B), the x-axis represents mRNA expression levels of transporters normalized to mRNA expression levels of villin 1, and the y-axis represents protein abundance. Correlation analysis was assessed using Spearman rank-order correlation (Rs) test with t-distribution of the P value. Linear regression analysis was carried out to assess the linearity of relationships and scatter of the data (R2). Correlations were deemed strong when the Rs values were greater than 0.60, P values were less than 0.05, and the data points demonstrated limited scatter (R2 > 0.30). Rs values between 0.5 and 0.6 with a P value less than 0.05 were considered to represent moderate correlation. Clear circles with a green border correspond to ileal samples, and clear circles with a red border correspond to jejunal samples.
Covariates of Expression of Intestinal Enzymes and Transporters.
Trends in protein expression of DMEs and transporters were assessed with reference to several demographic and clinical factors, including sex, age, and disease state of donors. Demographic and clinical information on donors is provided in Supplemental Table 1. There were no statistically significant differences in protein abundance of DMEs and transporters between male (n = 12) and female (n = 4) donors and very little difference between healthy donors (n = 5) and those with inflammatory bowel disease (n = 3), colon cancer (n = 5), or ischemia (n = 3). A weak correlation (R2 < 0.25 for all targets) was observed between protein expression and age (17–80 years).
Discussion
Drug absorption across the gut wall is a complex process. Drug bioavailability depends on the physicochemical properties of the drug and formulation and the activity of DMEs and drug transporters in the intestine (Olivares-Morales et al., 2015; Gao et al., 2017; Cristofoletti et al., 2018; Darwich et al., 2019). Regional differences in relative expression of enzymes and transporters can therefore play a significant role in defining the amount of drug that enters systemic circulation unchanged (Harwood et al., 2013; Drozdzik et al., 2014). Using a highly sensitive mass spectrometry platform and targeted quantitative proteomics (QconCAT) on human intestinal tissue from two different regions, jejunum and ileum, the abundance of various DMEs and drug transporters was quantified. Relative mRNA expression of several transporters was also measured. Although it is acknowledged that the duodenum could be a significant absorptive site for certain high-permeability drugs, duodenal tissues were not available for this study. Resection of duodenum is rare, and the surgical team associated with this study does not undertake procedures in this intestinal region of the bowel. However, the analysis of the jejunum and ileum regions of the bowel undertaken in this study is relevant because PBPK models generally consider the jejunum or ileum as the reference site for drug absorption/active transport because of their larger surface area and mean drug residence time (Bolger et al., 2009; Harwood et al., 2013).
All targeted P450 enzymes were expressed in both jejunum and ileum, with CYP3A4 being the most abundant P450, as previously reported (Paine et al., 2006; Drozdzik et al., 2018), indicating the primary importance of CYP3A4 in presystemic drug metabolism (Supplemental Table 5). In contrast with other studies in which CYP2D6 is frequently below the limit of quantification (Supplemental Table 5), CYP2D6 was quantified in all analyzed samples. In individuals with a poor metabolizer phenotype, the expression of this enzyme is known to be compromised (Dickinson et al., 2007). In the absence of related genotype information, which was not possible to obtain because of ethical constraints for this study, our results may indicate that the donors in the present study were not “poor metabolizers,” because CYP2D6 was quantified in all samples with little variability in the same range as CYP2C9, which is frequently reported as the second most abundant enzyme in intestine (Miyauchi et al., 2016; Drozdzik et al., 2018). Differences between reported abundances of DMEs can also be associated with interlaboratory differences in sample preparation methodologies. For example, some studies used intestinal microsomal fractions from enterocytes, others used total membrane-enriched fractions, and others used the entire crude mucosa containing other types of intestinal cells. Interlaboratory differences in the proteomic workflow can also contribute to technical variability in reported values (Harbourt et al., 2012, 2014; Sato et al., 2014; Nakamura et al., 2016; Akazawa et al., 2018).
UGT1A1 was the most abundant UGT, followed by UGT2B7 and UGT1A3, which is partially in agreement with published literature (Harbourt et al., 2012; Sato et al., 2014; Miyauchi et al., 2016; Nakamura et al., 2016). The concentration of UGT1A6 was equal in jejunum and ileum. Notably, we quantified UGT2B15 in both regions in spite of its low concentration and challenges associated with quantifying this enzyme. So far, UGT2B15 abundance has been reported in one individual within a population of 28 individuals (Miyauchi et al., 2016). In agreement with published data, the low abundance of these enzymes highlights the relatively minor role of these UGTs in comparison with P450 in intestinal drug clearance (Drozdzik et al., 2018).
A study in which ileum and jejunum samples were examined from the same donors reported differences in DME abundances between ileum and jejunum (Drozdzik et al., 2018). In our study, no significant differences were observed between the abundance of DMEs in jejunum and ileum except in the case of CYP3A4. This may be because the ileum and jejunum samples were not obtained from the same donor and because differences were obscured by high interindividual variability and the small number of jejunum samples analyzed.
In agreement with other studies, high intestinal abundances of Na+/K+-ATPase and cadherin-17 were quantified (Harwood et al., 2015, 2016). This highlights the applicability of total mucosal protein fractions to quantify both microsome-localized DMEs and plasma membrane marker proteins. MRP2 was found to be the most abundant ABC transporter, whereas OATP2B1 was the most abundant SLC transporter. Conflicting reports are found in the literature with respect to the rank order of abundances of the transporters quantified in this study (Gröer et al., 2013; Drozdzik et al., 2014, 2019; Miyauchi et al., 2016). This can be due to differences in sample preparation techniques and proteomic and interindividual variability (Gröer et al., 2013; Drozdzik et al., 2014; Harwood et al., 2014, 2016). Notably, we were able to quantify OATP1A2, which has not been detected in other studies (Hilgendorf et al., 2007; Meier et al., 2007; Drozdzik et al., 2014). The presence of OATP1A2 in the intestine has been disputed, although it has been observed immunohistochemically and is implicated in intestinal drug interactions with fruit juice (Glaeser et al., 2007; Tamai, 2012; Estudante et al., 2013). The high selectivity and sensitivity of our methodology provide confidence in our ability to detect this protein in these samples. Similar quantitative values of OST-α and OST-β are reported in our study, which conforms to our understanding that these subunits form a dimeric complex that is functionally expressed in the basal membrane of enterocytes (Seward et al., 2003).
Intercorrelations in protein abundance between DMEs and/or drug transporters have recently started to be used in PBPK models with the aim of achieving biologically plausible predictions of drug pharmacokinetics (Melillo et al., 2019). For instance, in studies involving the human liver, the utility of accounting for the intercorrelation between CYP3A4 and CYP2C8 abundance has been demonstrated in predicting drug clearance and drug-drug interactions between repaglinide and gemfibrozil (Doki et al., 2018). In a separate study involving human liver microsomes, strong intercorrelations have also been reported for several P450 and UGT enzymes; however, their physiologic relevance is unclear (Achour et al., 2014). As the functional interplay between DMEs and drug transporters is increasingly explored, the analysis of protein abundance intercorrelations presents valuable data for model refinement. Intercorrelations were found between P450, between UGTs, between P450 and UGTs, and between DMEs and transporters, in line with that previously established in studies using mRNA expression levels at the mRNA level; strong intercorrelations have been reported between CYP3A4-ABCB1 (P-gp) and UGT1A1-ABCC2 (MRP2) (Drozdzik et al., 2014; Bruckmueller et al., 2017; Fritz et al., 2019). In the present study, similar strong intercorrelation was observed at the protein level between these protein pairs. It is known that CYP3A4 and P-gp work synergistically, leading to an efficient first-pass drug extraction in the intestine (Cummins et al., 2002). There is also evidence associating the transmembrane transport of hydrophilic glucuronide metabolites formed by UGTs via MRP2 (Li et al., 2019); therefore, these correlations have some physiologic basis. A strong correlation observed between the P-gp and OST-α abundances in the gut wall (R2 = 0.77) is only expected if they are regulated by the same nuclear receptors and/or if they share a common regulatory pathway (Tanigawara et al., 1999; Kast et al., 2002; Luo et al., 2002; Trubetskoy et al., 2005; Urquhart et al., 2007). For many proteins studied here, the organ-specific regulatory mechanisms are likely to be complex and not consistently well characterized. In the present study, no significant correlation was observed between the P-gp and BCRP abundance despite a significant intercorrelation being found between their respective mRNA levels (P = 0.009, R2 = 0.31). The presence or absence of correlations between mRNA and protein has previously been described, and post-translational differences may play a role depending on age or disease-related protein modification and processing (Harwood et al., 2016).
There is a growing body of evidence that mRNA expression is a poor surrogate for protein abundance in the absence of steady-state conditions. This is relevant to the dynamic environment of the gut lumen. Environmental inducers (daily variations in food and beverages) and a high turnover of enterocytes [3.5 days (Darwich et al., 2014)] are likely to make the expression of proteins in the gut epithelium a dynamic process, making steady-state conditions unlikely, thus justifying the use of protein abundance in preference to mRNA in PBPK models extrapolated in vitro to in vivo.
Various demographic covariates were assessed for their effects on protein abundance, including gender and age. There was no significant relationship with protein expression levels in either region of the intestine. However, the value of the covariate analysis is limited because of the small sample size. Covariate analysis has not been undertaken before mainly because of the low number of samples used in previous studies (Oswald et al., 2013; Drozdzik et al., 2014; Harwood et al., 2015). A more recent study (Miyauchi et al., 2016) assessed the effects of sex, age, smoking, and obesity on the expression of enzymes and transporters in the jejunal membrane fractions from 28 donors with morbid obesity, and there was an overall lack of effect of age or gender on expression. These observations are consistent with data from the liver, which showed limited differences due to gender and lack of correlation with age after maturity (Achour et al., 2014; Prasad et al., 2014; Couto et al., 2019). The impact of drugs administered to individuals in this study should also be carefully considered. The proton pump inhibitor omeprazole, which is predominantly metabolized by CYP2C19, has been shown to downregulate CYP2C19 expression (Zvyaga et al., 2012). In our study, 7 out of 16 donors were administered omeprazole, but no effect on CYP2C19 expression was observed.
It is intended that the data generated in this study will be incorporated into whole-body PBPK models. However, the matrix in which the abundances of drug-metabolizing enzymes and drug transporters were measured requires careful consideration and application of appropriate scaling factors before incorporation into the PBPK model. Ongoing efforts in our research group are focused on developing appropriate scaling factors.
In conclusion, the present study provides an in-depth analysis of protein abundance and protein abundance intercorrelations between DMEs and transporters in the human jejunum and ileum. Various covariates of expression were examined, and expression of pharmacologically active proteins was shown to be independent of gender and age. The use of quantitative measurements of protein abundance together with other physiologic and drug-specific measurements can facilitate predictions of drug absorption and clearance in the human intestine. The data in this study contribute to a growing body of evidence reporting absolute enzyme and transporter abundances in the human intestine that can be incorporated into population-based PBPK models.
Acknowledgments
The authors thank the Bio-MS core facility, University of Manchester, for providing access to Liquid Chromatography–Mass Spectrometry instrumentation and assistance with method development. The authors also thank Sibylle Neuhoff for constructive comments during preparation of this manuscript and Eleanor Savill for helping to format and submit the manuscript.
Authorship Contributions
Participated in research design: Harwood, Carlson, Rostami-Hodjegan, Warhurst.
Conducted experiments: Couto, Al-Majdoub, Gibson, Davies, Achour, Harwood.
Contributed new reagents or analytic tools: Couto, Al-Majdoub.
Performed data analysis: Couto, Al-Majdoub, Achour, Barber.
Wrote or contributed to the writing of the manuscript: Couto, Al-Majdoub, Gibson, Davies, Achour, Harwood, Carlson, Barber, Rostami-Hodjegan, Warhurst.
Footnotes
- Received October 16, 2019.
- Accepted December 23, 2019.
This work was supported by the Innovative Medicines Initiative Joint Undertaking (http://www.imi.europa.eu) [Grant Agreement 115369], resources from which are composed of financial contributions from the European Union’s Seventh Framework Programme [FP7/2007-2013] and The European Federation of Pharmaceutical Industries and Associations companies’ in-kind contribution. An award from the Royal Commission for the Exhibition of 1851 to the University of Manchester based on MDH successfully completing an Industrial Fellowship grant in 2015 facilitated the initial aspects of this work.
↵
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
Abbreviations
- BCRP
- breast cancer resistance protein
- MRP2
- multidrug resistance-associated protein 2
- OST-α
- organic solute transporter subunit alpha
- OST-β
- organic solute transporter subunit beta
- OATP2B1
- organic anion transporter polypeptide-related protein 2
- OATP1A2
- organic anion-transporting polypeptide 1
- P-gp
- P-glycoprotein 1
- Na+/K+
- ATPase sodium/potassium-transporting ATPase subunit alpha-1
- HPLC
- high-performance liquid chromatography
- ABC ATP
- binding cassette
- CDH-17
- cadherin-17
- DME
- drug-metabolizing enzyme
- GAPDH
- glyceraldehyde 3-phosphate dehydrogenase
- MetCAT
- concatemer of standard peptides from human drug metabolizing enzymes
- MRM
- multiple reaction monitoring
- P450
- cytochrome P450
- PBPK
- physiologically based pharmacokinetic
- QconCAT
- quantification concatemer
- RIPA
- radioimmunoprecipitation assay
- Rs
- Spearman rank-order correlation
- SLC
- solute carrier
- TransCAT
- concatemer of standard peptides from human hepatic transporters
- UGT
- uridine 5′-diphosphate-glucuronosyltransferase
- UPL
- Universal Probe Library
- Copyright © 2020 by The Author(s)
This is an open access article distributed under the CC BY-NC Attribution 4.0 International license.