Abstract
Flutamide, an antiandrogen drug, is widely used for the treatment of prostate cancer. The initial metabolic pathways of flutamide are hydroxylation and hydrolysis. It was recently reported that the hydrolyzed product, 4-nitro-3-(trifluoromethyl)phenylamine (FLU-1), is further metabolized to N-hydroxy FLU-1, an assumed hepatotoxicant. However, the esterase responsible for the flutamide hydrolysis has not been characterized. In the present study, we found that human arylacetamide deacetylase (AADAC) efficiently hydrolyzed flutamide using recombinant AADAC expressed in COS7 cells. In contrast, carboxylesterase1 (CES1) and CES2, which are responsible for the hydrolysis of many drugs, could not hydrolyze flutamide. AADAC is specifically expressed in the endoplasmic reticulum. Flutamide hydrolase activity was highly detected in human liver microsomes (Km, 794 ± 83 μM; Vmax, 1.1 ± 0.0 nmol/min/mg protein), whereas the activity was extremely low in human liver cytosol. The flutamide hydrolase activity in human liver microsomes was strongly inhibited by bis-(nonylphenyl)-phenylphosphate, diisopropylphosphorofluoride, and physostigmine sulfate (eserine) but moderately inhibited by sodium fluoride, phenylmethylsulfonyl fluoride, and disulfiram. The same inhibition pattern was obtained with the recombinant AADAC. Moreover, human liver and jejunum microsomes showing AADAC expression could hydrolyze flutamide, but human pulmonary and renal microsomes, which do not express AADAC, showed slight activity. In human liver microsomal samples (n = 50), the flutamide hydrolase activities were significantly correlated with the expression levels of AADAC protein (r = 0.66, p < 0.001). In conclusion, these results clearly showed that flutamide is exclusively hydrolyzed by AADAC. AADAC would be an important enzyme responsible for flutamide-induced hepatotoxicity.
Flutamide (3′-trifluoro-2-methyl-4′-nitro-2-methyl-propinoylanilide) is a nonsteroidal antiandrogen drug used for the treatment of prostate cancer. The combination of luteinizing hormone-releasing hormone agonist results in prolonged survival in prostate cancer patients (Crawford et al., 1989). However, flutamide occasionally causes severe hepatotoxicity (Thole et al., 2004). Flutamide itself is not toxic when used at the appropriate clinical dose, but bioactivation of flutamide has been considered to be the cause of flutamide-induced hepatotoxicity (Fau et al., 1994).
Flutamide is mainly metabolized to 2-hydroxyflutamide by human CYP1A2. It has been suggested that 2-hydroxyflutamide is associated with the therapeutic effect of flutamide (Katchen and Buxbaum, 1975). Flutamide is also hydrolyzed to 4-nitro-3-(trifluoromethyl)phenylamine (FLU-1) by esterase (Katchen and Buxbaum, 1975; Schulz et al., 1988). FLU-1 is considered to have no therapeutic effect (Aizawa et al., 2003). Goda et al. (2006) recently reported that FLU-1 is further metabolized to N-hydroxyl FLU-1 by human CYP3A4. Many researchers have reported on the relationship between the toxicity and metabolism of flutamide. It was shown in CYP1A2 knockout SV129 mice but not in wild-type mice that the urinary concentration of FLU-1 was increased, and an abnormal elevation of alanine aminotransferase was shown after the knockout mice were fed an amino acid-deficient diet (Matsuzaki et al., 2006). It was also shown in humans that the urinary caffeine metabolite ratio, an indicator of the CYP1A2 activity, was significantly lower in patients with hepatic injury compared with patients with normal hepatic function after the same flutamide therapy (Ozono et al., 2002). In addition, a study on patients of prostate cancer taking flutamide showed that the incidence of hepatotoxicity was correlated with the plasma concentration of FLU-1 (Aizawa et al., 2003). More recently, Ohbuchi et al. (2009) reported that coadministration of FLU-1 and 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene, an inducer of CYP3A and CYP1A, to mice significantly increased serum alanine aminotransferase, and several protein adducts were detected after incubation of the microsomal protein with N-hydroxy FLU-1. Therefore, the hepatotoxicity of flutamide might be related to N-hydroxy FLU-1. Considering these reports, the hydrolysis pathway may contribute to the hepatotoxicity of flutamide.
Carboxylesterase (CES) is the major serine esterase contributing to the hydrolysis of various drugs and xenobiotics. In human, CES isoforms are classified into three families: CES1, CES2, and CES3. CES1 and CES2 have been reported to be responsible for the biotransformation of a number of clinically used drugs and prodrugs such as imidapril, capecitabine, and irinotecan (Imai et al., 2006). CES3 appears to show extremely low activity compared with CES1 and CES2 (Sanghani et al., 2004). The flutamide hydrolysis by human liver microsomes (HLM) is inhibited by bis-(nonylphenyl)-phenylphosphate (BNPP), a general CES inhibitor (Heymann and Krisch, 1967; Block and Arndt, 1978; Mentlein et al., 1988). Thus, it is plausible that flutamide is hydrolyzed by CES. However, the flutamide hydrolase activity was not detected by using purified CES1 (pI 4.5) and CES2 (pI 5.3) (Takai et al., 1997). Therefore, we considered that another esterase expressed in HLM plays a role in flutamide hydrolysis. As a candidate enzyme, arylacetamide deacetylase (AADAC) could be considered.
AADAC, as well as CES1 and CES2, is a major serine hydrolase expressed in HLM (Ross and Crow, 2007). AADAC was first identified as the enzyme that catalyzes the deacetylation of 2-acetylaminofluorene (Probst et al., 1991). The active site domain of AADAC shares high homology with that of hormone-sensitive lipase (Probst et al., 1994). Therefore, AADAC has been classified as a lipase. In fact, Tiwari et al. (2007) proved that human AADAC was capable of hydrolyzing cholesterol ester when expressed in yeast. However, it is unknown whether AADAC is involved in the hydrolysis of clinical therapeutic drugs. In the present study, the involvement of human AADAC in flutamide hydrolysis was investigated.
Materials and Methods
Chemicals and Reagents. Flutamide, 4-nitro-3-(trifluoromethyl)phenylamine (FLU-1), p-nitrophenol, diisopropylphosphorofluoride (DFP), physostigmine sulfate (eserine), phenylmethylsulfonyl fluoride (PMSF), disulfiram, silver nitrate (AgNO3), cadmium chloride (CdCl2), cobaltous chloride (CoCl2), and cupric chloride (CuCl2) were purchased from Wako Pure Chemical Industries (Osaka, Japan). p-Nitrophenyl acetate (PNPA), BNPP, and sodium fluoride (NaF) were purchased from Sigma-Aldrich (St. Louis, MO). Primers were commercially synthesized at Hokkaido System Sciences (Sapporo, Japan). The random hexamer and SYBR Premix Ex Taq were from Takara (Shiga, Japan). RevaTra Ace (Moloney murine leukemia virus reverse transcriptase RNase H–) was obtained from Toyobo (Tokyo, Japan). All the other chemicals used in this study were of analytical or the highest quality commercially available.
Human Tissues. The microsomes or cytosol from human liver, jejunum, lung, and kidney were used in this study. Pooled HLM (n = 50), individual HLM (24 donors), and pooled human liver cytosol (HLC, n = 22) were purchased from BD Gentest (Woburn, MA). Individual HLM from 15 donors were also obtained from Human and Animal Bridging Research Organization (Chiba, Japan), and those from 11 donors were obtained from autopsy materials that were discarded after pathological investigation. Human jejunum microsomes (HJM, pooled, n = 10), human pulmonary microsomes (HPM, single donor), and human renal microsomes (HRM, single donor) were purchased from Tissue Transformation Technologies (Edison, NJ). The pooled HLM, HJM, HPM, and HRM were used for the immunoblotting analysis and the assay of flutamide hydrolase activity. The HLC was used for the comparison of flutamide hydrolase activity with HLM. The individual HLM samples were used for the correlation analysis. The use of the human tissues was approved by the Ethics Committees of Kanazawa University (Kanazawa, Japan) and Iwate Medical University (Morioka, Japan).
Total RNA from Human Tissues and Reverse Transcription-Polymerase Chain Reaction Analyses. Total RNA samples from normal human liver (single donor), colon (pooled, n = 2), kidney (single donor), bladder (pooled, n = 2), breast (pooled, n = 2), ovary (single donor), and uterus (pooled, n = 3) were obtained from Stratagene (La Jolla, CA). Total RNA samples from normal human lung (single donor) and testis (single donor) were from Cell Applications (San Diego, CA). Total RNA samples from normal human stomach (single donor), adrenal gland (pooled, n = 62), and small intestine (pooled, n = 5) were from Clontech (Palo Alto, CA). The reverse transcription procedure was described previously (Nakajima et al., 2006).
For quantitative analysis, real-time reverse transcription-polymerase chain reaction (RT-PCR) was performed for AADAC mRNA using an MX3000P real-time PCR system (Stratagene). The forward and reverse primers used for PCR were AADAC-RT-S and AADAC-RT-AS primers (Table 1). A 1-μl portion of the reverse-transcribed mixture was added to a PCR mixture containing 10 pmol of each primer and 12.5 μl of SYBR Premix Ex Taq solution in a final volume of 25 μl. After an initial denaturation at 95°C for 30 s, the amplification was performed by denaturation at 94°C for 4 s, annealing at 58°C for 7 s, and extension at 72°C for 20 s for 45 cycles. Human glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA was also quantified according to a method described previously (Tsuchiya et al., 2004). The copy numbers were calculated using standard amplification curves.
Primers used in the present study
Construction of Plasmids Expressing Human AADAC, CES1A, and CES2. The full-length human AADAC, CES1A1, CES1A2, and CES2A1 cDNAs were obtained by RT-PCR using a human liver RNA sample as the initial template. The primers used are shown in Table 1. In this study, two clones of AADAC cDNA were obtained [c.931guanine (G) and adenine (A)]. In the reference sequence of NM 001086.2, the nucleotide at the c.931 position is G. Therefore, the clones with c.931G and c.931A were defined as the AADAC wild-type and AADAC variant, respectively. This single nucleotide polymorphism (SNP, c.931G>A) leads to an amino acid change of valine to isoleucine. The allele frequency of this SNP (ID: rs1803155) has been reported to be approximately 55 to 80% in each population, including European, Asian, and sub-Saharan African in the dbSNP database in the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/SNP/snp_ref.cgi?rs=1803155). The PCR products were subcloned into pTARGET mammalian expression vector (Promega, Madison, WI). The nucleotide sequences were confirmed by DNA sequence analysis (Long-Read Tower DNA sequencer; GE Healthcare, Little Chalfont, Buckinghamshire, UK).
Transfection of Plasmids Expressing Human AADAC, CES1A, and CES2. African green monkey kidney cells, COS7 cells, were obtained from American Type Culture Collection (Manassas, VA). The COS7 cells were grown in Dulbecco's modified Eagle's medium containing 4.5 g/l glucose and 10% fetal bovine serum with 5% CO2 at 37°C. The cells were transfected in 10-cm dishes with 7.5 μg of each expression plasmid using Lipofectamine (Invitrogen, Carlsbad, CA). After incubation for 48 h, the cells were harvested and suspended in a small amount of TGE buffer (10 mM Tris-HCl, 20% glycerol, 1 mM EDTA, pH 7.4) and disrupted by freeze-thawing three times. Each protein expression level was determined by immunoblot analysis as described below.
Immunoblot Analysis. SDS-polyacrylamide gel electrophoresis and immunoblot analysis were performed according to Laemmli (1970). Enzyme sources (30 μg) were separated on 10% polyacrylamide gels and electrotransferred onto polyvinylidene difluoride membrane, Immobilon-P (Millipore Corporation, Billerica, MA). The membranes were probed with monoclonal mouse anti-human AADAC (Abnova, Neihu District, Taipei City, Taiwan), polyclonal rabbit anti-human CES1 (Abcam, Cambridge, MA), and polyclonal rabbit anti-human CES2 antibodies (Antagene, San Francisco, CA), and the corresponding fluorescent dye-conjugated second antibody and an Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE) were used for the detection. The relative expression level was quantified using ImageQuant TL Image Analysis software (GE Healthcare).
Flutamide Hydrolase Activity. The flutamide hydrolase activity was determined as follows: a typical incubation mixture (final volume of 0.2 ml) contained 100 mM potassium phosphate buffer, pH 7.4, and various enzyme sources (human microsomal protein and COS7 cell homogenate expressing esterases, 0.4 mg/ml; human cytosolic protein, 1.0 mg/ml). In the preliminary study, we confirmed that the rate of formation of FLU-1 was linear with respect to the protein concentrations (<1.0 mg/ml human microsomal protein and COS7 cells homogenate expressing esterases and <1.5 mg/ml human cytosolic protein) and incubation time (<60 min). Flutamide was dissolved in dimethyl sulfoxide (DMSO), and the final concentration of DMSO in the incubation mixture was 1.0%. The reaction was initiated by the addition of 25 to 750 μM flutamide after 2-min preincubation at 37°C. After the 30-min incubation at 37°C, the reaction was terminated by the addition of 0.1 ml of ice-cold acetonitrile. After removal of the protein by centrifugation at 9500g for 5 min, a 60-μl portion of the supernatant was subjected to high-performance liquid chromatography (HPLC). The HPLC analysis was performed using an L-7100 pump (Hitachi, Tokyo, Japan), an L-7200 autosampler (Hitachi), an L-7405 UV detector (Hitachi), and a D-2500 Chromato-Integrator (Hitachi) equipped with a Mightysil RP-18 C18 GP column (5-μm particle size, 4.6 mm i.d.×150 mm; Kanto Chemical, Tokyo, Japan). The eluent was monitored at 376 nm with a noise-base clean Uni-3 (Union, Gunma, Japan), which can reduce the noise by integrating the output and increase the signal 3-fold by differentiating the output, and 5-fold by further amplification with an internal amplifier, resulting in a maximum 15-fold amplification of the signal. The mobile phase was 45% acetonitrile containing 25 mM ammonium acetate, pH 5.0. The flow rate was 1.0 ml/min. The column temperature was 35°C. The quantification of FLU-1 was performed by comparing the HPLC peak height with that of an authentic standard. Because FLU-1 contaminants exist in the commercially available flutamide to the extent of ∼0.5%, the content of FLU-1 in the mixture incubated without the enzyme was subtracted from that with the enzyme to correct the activity. The activity in each concentration was determined as the mean value in triplicate. For kinetic analyses of flutamide hydrolase activity, the parameters were estimated from the fitted curves using a computer program (KaleidaGraph, Synergy Software, Reading, PA) designed for nonlinear regression analysis.
To clarify the involvement of various esterases, inhibition analysis of flutamide hydrolysis was performed by using representative esterase inhibitors. Organophosphates such as BNPP and DFP are known as general CES inhibitors (Heymann and Krisch, 1967; Yamaori et al., 2006). Eserine and NaF are cholinesterase inhibitors (Iwatsubo, 1965; Preuss and Svensson, 1996). EDTA is a paraoxonase inhibitor (Gonzalvo et al., 1997). PMSF is a serine hydrolase inhibitor (Johnson and Moore, 2000). Because heavy metals are frequently used for esterase inhibition studies, AgNO3, CdCl2, CoCl2, and CuCl2 were also used as inhibitors. Disulfiram was reported as a monoacylglycerol lipase inhibitor (Labar et al., 2007). The concentration of BNPP and DFP ranged from 0.001 to 1 mM, and that of the others was 0.1 and 1 mM. PMSF and disulfiram were dissolved in DMSO such that the final concentration of DMSO in the incubation mixture was 1.5%. Other inhibitors were dissolved in distilled water. The experimental procedure and condition were the same as above except that 500 μM flutamide was added. It was confirmed that 1.5% DMSO did not inhibit the flutamide hydrolase activity, and the control activity was determined in the presence of 1.5% DMSO.
p-Nitrophenyl Acetate Hydrolase Activity. To confirm whether the recombinant AADAC, CES1, and CES2 are enzymatically active, the PNPA, a general esterase substrate, hydrolase activity was measured. The PNPA hydrolase activity was determined as follows: a typical incubation mixture was the same as above. PNPA was dissolved in DMSO, and the final concentration of DMSO in the incubation mixture was 1.0%. The reaction was initiated by the addition of 500 μM PNPA after 2-min preincubation at 37°C. After 5-min incubation at 37°C, the reaction was terminated by the addition of 0.1 ml of ice-cold methanol. The PNPA hydrolase activity was measured by the absorbance at 405 nm using Biotrak II plate reader (GE Healthcare). The quantification of p-nitrophenol, a metabolite of PNPA hydrolysis, was performed by comparing the absorbance with that of an authentic standard. Because p-nitrophenol contaminants exist in the commercially available PNPA to the extent of ∼5%, the content of p-nitrophenol in the mixture incubated without the enzyme was subtracted from that with the enzyme to correct the activity.
Statistical Analysis. Comparison of two groups was made with unpaired, two-tailed Student's t test. Correlation analyses were performed by Spearman rank method. A value of p < 0.05 was considered statistically significant.
Results
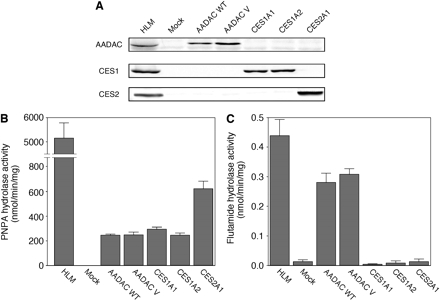
PNPA and Flutamide Hydrolase Activities by Recombinant Human AADAC Wild-Type, AADAC Variant, CES1A1, CES1A2, and CES2. To compare the flutamide hydrolase activity between human AADAC, CES1A1, CES1A2, and CES2, they were transiently expressed in COS7 cells. The AADAC variant was also transiently expressed in COS7 cells. The protein expression levels were determined by immunoblot analysis (Fig. 1A). AADAC protein was specifically expressed in COS7 cells transfected with the expression plasmids of AADAC wild-type and variant. CES1A and CES2 proteins were specifically expressed in COS7 cells transfected with the expression plasmids of CES1A1 or CES1A2, and CES2, respectively. To confirm that these enzymes were enzymatically active, PNPA hydrolase activity was measured at a concentration of 500 μM PNPA (Fig. 1B). The recombinant CES2A1 showed the highest PNPA hydrolase activity (621 ± 61 nmol/min/mg protein), and the recombinant AADAC wild-type, variant, CES1A1, and CES1A2 showed similar activities (243 ± 11, 247 ± 24, 293 ± 19, and 245 ± 18 nmol/min/mg protein, respectively). The flutamide hydrolase activities were determined at a concentration of 500 μM flutamide (Fig. 1C). Among them, the AADAC wild-type and variant showed flutamide hydrolase activity (0.28 ± 0.03 and 0.30 ± 0.01 nmol/min/mg protein, respectively). In contrast, CES1A1, CES1A2, and CES2 showed almost no activity (0.003 ± 0.003, 0.008 ± 0.008, and 0.012 ± 0.007 nmol/min/mg protein, respectively), similar to mock COS7 cells (0.012 ± 0.007 nmol/min/mg protein). These results suggested that AADAC, but not the CES enzymes, contributed to the flutamide hydrolysis.
Kinetic Analyses of Flutamide Hydrolase Activity by HLM, HLC, and Recombinant Human AADAC. It was previously suggested that AADAC is localized to the endoplasmic reticulum lumen (Frick et al., 2004). Therefore, it is assumed that flutamide can be hydrolyzed in HLM rather than in HLC. In this study, the flutamide hydrolase activity in HLM and HLC was measured (Fig. 2A). The maximum concentration was 750 μM because of the limited solubility of flutamide in the incubation mixture. For HLM, the Km and Vmax values were 794 ± 83 μM and 1.1 ± 0.0 nmol/min/mg protein, respectively, resulting in an intrinsic clearance of 1.4 ± 0.1 μl/min/mg protein. For HLC, because the flutamide hydrolase activity ranging from 25 to 750 μM was linear, the Km and Vmax values could not be calculated by the Michaelis-Menten equation. The Km value of the flutamide hydrolase activity by HLC appeared to be substantially higher than that by HLM. These results suggested that the contribution to flutamide hydrolysis by HLM was much higher than that by HLC. In addition, kinetic analyses of the flutamide hydrolase activity by the recombinant AADAC wild-type and variant were also performed (Fig. 2B). The Km and Vmax values of the AADAC wild-type were 778 ± 122 μM and 0.6 ± 0.1 nmol/min/mg protein, respectively, resulting in an intrinsic clearance of 0.8 ± 0.0 μl/min/mg protein. The Km and Vmax values of the AADAC variant were 591 ± 75 μM and 0.5 ± 0.0 nmol/min/mg protein, respectively, resulting in an intrinsic clearance of 0.9 ± 0.1 μl/min/mg protein. Thus, the AADAC variant did not alter the flutamide hydrolase activity. In addition, the Km values of HLM and AADAC wild-type were not significantly different. These results suggested that AADAC was involved in flutamide hydrolysis in human liver.
A, immunoblot analysis of recombinant human AADAC, CES1, and CES2 expressed in COS7 cells. Total cell homogenates from COS7 cells (30 μg) were separated by electrophoresis using 10% SDS-polyacrylamide gel. PNPA hydrolase activities (B) and flutamide hydrolase activities (C) by recombinant AADAC, CES1, and CES2. The homogenates of COS7 cells expressing these enzymes were incubated with 100 μM PNPA or 500 μM flutamide. Each column represents the mean ± S.D. of triplicate determinations. WT, wild type; V, variant.
Kinetic analyses of flutamide hydrolase activities in HLM and HLC (A) and by the recombinant AADAC wild-type and variant (B). The kinetic parameters were estimated from the fitted curve using the computer program KaleidaGraph designed for nonlinear regression analysis. Each data point represents the mean ± S.D. of triplicate determination.
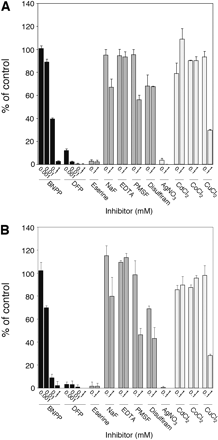
Effects of Chemical Inhibitors on Flutamide Hydrolase Activity in HLM and Recombinant AADAC. To prove that AADAC is a principal enzyme for flutamide hydrolysis in human liver, the effects of inhibitors on the flutamide hydrolase activities by HLM and the recombinant AADAC were analyzed. In this study, representative inhibitors of various esterases were used to clarify the involvement of various esterases and examine the inhibitory characteristics of AADAC. The flutamide hydrolase activity by HLM was inhibited in a BNPP concentration-dependent manner and was potently inhibited by 0.01 to 1 mM DFP, 0.1 to 1 mM eserine, and AgNO3 (Fig. 3A). In addition, the flutamide hydrolase activity by HLM was moderately inhibited by 1 mM NaF, PMSF, disulfiram, and CuCl2. However, no inhibition occurred by EDTA, CaCl2, and CoCl2. A similar inhibition pattern was obtained by the recombinant AADAC wild-type (Fig. 3B) and variant (data not shown). These results imply that AADAC is a major esterase responsible for the flutamide hydrolysis in human liver.
Effects of chemical inhibitors on flutamide hydrolase activity. Flutamide hydrolase activities in HLM (A) and the recombinant AADAC (B) wild-type were determined at a substrate concentration of 500 μM. The control activity values were 0.438 (A) and 0.283 (B) nmol/min/mg protein, respectively. Each column represents the mean ± S.D. of triplicate determinations.
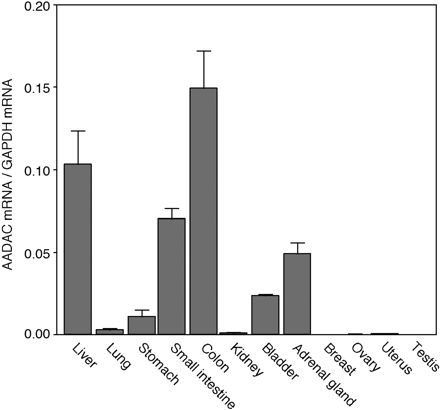
Expression of AADAC mRNA in Human Normal Tissues. The expression level of AADAC mRNA in human tissues was determined by real-time RT-PCR analysis (Fig. 4). The expression level of AADAC mRNA was normalized with GAPDH mRNA level. AADAC mRNA is highly expressed in colon, liver, and small intestine and moderately expressed in adrenal gland, bladder, and stomach. Other tissues investigated in this study showed low expression levels. We previously examined the expressions of CES1A1, CES1A2, and CES2 mRNA in human normal tissues (T. Maruichi, M. Katoh, S. Takahashi, M. Nakajima, and T. Yokoi, unpublished data). The expression levels of AADAC mRNA in all the tissues except colon and adrenal gland were lower than those of these CES mRNA. The expression level of AADAC in human liver was 116-, 23-, and 8-fold lower than those of CES1A1, CES1A2, and CES2, respectively.
Expression levels of AADAC mRNA in various human normal tissues. Relative copy numbers of AADAC to GAPDH in human tissues were determined by real-time RT-PCR analysis. Each column represents the mean ± S.D. of triplicate determinations.
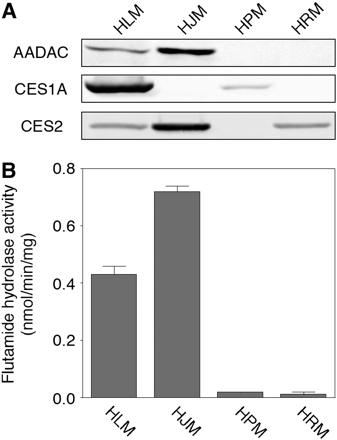
Expression of AADAC Protein and Flutamide Hydrolase Activities in Human Tissues. To further analyze whether AADAC is responsible for the flutamide hydrolysis in humans, the expression of AADAC and the flutamide hydrolase activity in various human tissues were measured. The expression levels of AADAC, CES1A, and CES2 proteins in HLM, HJM, HPM, and HRM were determined by immunoblot analysis (Fig. 5A). AADAC protein was expressed in HLM and HJM but not in HPM and HRM. This result corresponded with the expression of AADAC mRNA (Fig. 4). CES1A protein was expressed in HLM and HPM, whereas CES2 protein was expressed in HLM, HJM, and HRM. High flutamide hydrolase activity at a concentration of 500 μM flutamide was detected in HJM (0.72 ± 0.02 nmol/min/mg protein) and HLM (0.43 ± 0.03 nmol/min/mg protein). The fact that the hydrolase activity in HJM was higher than that in HLM was supported by the results of the immunoblot analysis. On the other hand, HPM, in which CES1A is expressed, and HRM, in which CES2 is expressed, showed slight hydrolase activity (0.02 ± 0.00 and 0.01 ± 0.01 nmol/min/mg protein, respectively). These results suggested that AADAC is responsible for the flutamide hydrolysis in humans.
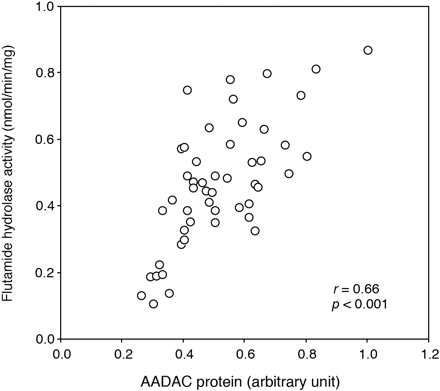
Correlation Analysis between Flutamide Hydrolase Activity and AADAC Protein Expression Level. The flutamide hydrolase activities in microsomes from 50 human livers were determined at a concentration of 500 μM. The flutamide hydrolase activities ranged from 0.11 to 0.87 nmol/min/mg protein (mean ± S.D., 0.47 ± 0.19 nmol/min/mg protein), resulting in 7.9-fold interindividual variability. In addition, the expression levels of AADAC protein in microsomes from 50 human livers were determined by immunoblot analysis. The AADAC protein expression levels are represented as relative levels to the sample, which is the highest expression level. The relative expression levels of AADAC protein in HLM ranged from 0.26 to 1.00, resulting in 3.8-fold interindividual variability. As shown in Fig. 6, the expression level of AADAC protein and the flutamide hydrolase activity were significantly correlated (r = 0.66, p < 0.001). These results also supported that AADAC is a principal enzyme for the flutamide hydrolysis.
A, immunoblot analysis of AADAC, CES1A, and CES2 in HLM, HJM, HPM, and HRM. Total cell homogenates from COS7 cells (30 μg) were separated by electrophoresis using 10% SDS-polyacrylamide gel. B, flutamide hydrolase activities in microsomes. The microsomes were incubated with 500 μM flutamide. Each column represents the mean ± S.D. of triplicate determinations.
Discussion
Human AADAC was first identified as an enzyme that catalyzes the deacetylation of 2-acetylaminofluorene (Probst et al., 1991). It has been believed that AADAC might function as a lipase because of the high homology of the active site of AADAC with that of hormonesensitive lipase (Probst et al., 1994). On the other hand, human CES enzymes are major serine esterases involved in the hydrolysis of various drugs and xenobiotics. AADAC is one of the major serine esterases expressed in HLM and CES enzymes (Ross and Crow, 2007), but it was unknown whether AADAC is involved in the hydrolysis of therapeutic drugs.
An antiandrogen drug, flutamide, has been widely used for prostate cancer, but severe hepatotoxicity sometimes occurred. Several studies suggested that the flutamide-induced hepatotoxicity was caused by FLU-1, a metabolite of hydrolyzed flutamide, or N-hydroxyl FLU-1, a metabolite of FLU-1 by CYP3A or CYP1A (Aizawa et al., 2003; Matsuzaki et al., 2006; Ohbuchi et al., 2009). Therefore, it was considered that flutamide hydrolysis was important in the occurrence of hepatotoxicity, but the flutamide hydrolase enzyme had never been identified. It was conceivable that human CES enzymes CES1A and CES2 are responsible for the hydrolysis of flutamide because they contribute largely to the hydrolysis of various drugs and xenobiotics. However, it was reported that the flutamide hydrolase activity was not detected by using purified CES1A and CES2 (Takai et al., 1997), which is consistent with our finding that the recombinant CES1A1, CES1A2, and CES2A1 did not show the flutamide hydrolase activity (Fig. 1C). Moreover, the activity was scarcely detected in HPM and HRM, in which CES1A and CES2 enzymes were expressed, respectively. On the other hand, the recombinant AADAC showed the flutamide hydrolase activity, and the activity was highly detected in HLM and HJM, in which AADAC is expressed. Furthermore, the similar activities of flutamide hydrolysis in HLM and the recombinant AADAC were consistent with the similar blot densities of AADAC (Fig. 1, A and C). Thus, we found that human AADAC is a major contributor to the flutamide hydrolysis. PNPA hydrolase activity, a general esterase activity, was detected in recombinant AADAC and CES1A1, CES1A2, and CES2A1 (Fig. 1B). However, the activity in HLM was substantially higher than the sum of those by recombinant AADAC and CES enzymes (Fig. 1B). Other esterases such as paraoxonase and butyrylcholinesterase may participate in the PNPA hydrolysis in HLM.
Correlation between the expression levels of AADAC protein and flutamide hydrolase activities in 50 HLM. The expression level of AADAC protein was determined by immunoblot analysis. The flutamide hydrolase activity was determined by HPLC.
It has been reported that AADAC is located on the lumen side of endoplasmic reticulum (Frick et al., 2004). This corresponded to the present result that the flutamide hydrolase activity in HLM was substantially higher than in HLC (Fig. 2A). In addition, there was no significant difference between the Km values of HLM (794 ± 83 μM) and recombinant AADAC wild-type (778 ± 122 μM) (Fig. 2, A and B). This finding also supported the involvement of AADAC in flutamide hydrolysis in human liver. However, the activity was also detected in HLC. It was already known that CES1A and CES2 enzymes are present in HLC (Xu et al., 2002; Tabata et al., 2004), but recombinant CES1A1, CES1A2, and CES2 could not hydrolyze flutamide (Fig. 1C). In addition, it was shown that sialic acid 9-O-acetylesterase, an alacepril hydrolase enzyme, purified from rat liver cytosol also could not hydrolyze flutamide (Usui et al., 2003). Therefore, other enzymes that can hydrolyze flutamide may be present in HLC, but the contribution to flutamide hydrolysis would be limited.
To confirm that flutamide hydrolysis in HLM is specifically catalyzed by AADAC, we performed inhibition analyses by using various chemical inhibitors and heavy metals (Fig. 3). The flutamide hydrolase activity in HLM was inhibited in a BNPP concentration-dependent manner but was not completely inhibited by 1 mM PMSF (residual activity, 56% of control). It is known that the CES1A and CES2 enzyme activities are inhibited in a BNPP concentration-dependent manner and were substantially inhibited by 0.1 mM PMSF (Xie et al., 2002). Therefore, these enzymes would not contribute to the flutamide hydrolysis in HLM. Moreover, the activity was potently inhibited by DFP, eserine, and silver nitrate and moderately inhibited by NaF, disulfiram, and CuCl2. Other inhibitors used in the present study did not cause inhibition. Because DFP and BNPP are organophosphates, general CES inhibitors, it is plausible that DFP also inhibits AADAC. Eserine and NaF are cholinesterase inhibitors (Iwatsubo, 1965; Ciliv and Ozand, 1972; Preuss and Svensson, 1996). It is of interest that flutamide hydrolysis was effectively inhibited by eserine but not by NaF. EDTA and disulfiram are known to be inhibitors of paraoxonase and monoacylglycerol lipase, respectively (Gonzalvo et al., 1997; Labar et al., 2007). Therefore, it was conceivable that these enzymes did not contribute to the flutamide hydrolysis in HLM. The inhibitory specificity of esterases by heavy metals has not been obvious, but in this study we found that AADAC was potently inhibited by AgNO3 and moderately inhibited by CuCl2. Probst et al. (1994) previously reported that the 2-acetylaminofluorene deacetylation catalyzed by AADAC in HLM was inhibited in a BNPP concentration-dependent manner but was not inhibited by increasing the concentrations of PMSF. The inhibition pattern of flutamide hydrolysis by BNPP and PMSF was similar to that of 2-acetylaminofluorene deacetylation.
To investigate the tissue distribution of AADAC mRNA in human normal tissues, real-time RT-PCR was performed (Fig. 4). AADAC mRNA was mainly expressed in human normal liver, small intestine, and colon. In addition, we confirmed that AADAC protein was expressed in human liver and jejunum microsomes but not in human pulmonary and renal microsomes (Fig. 5A). The tissue distribution of AADAC protein corresponded to that of AADAC mRNA. It is of interest that the expression of AADAC protein in jejunum was higher than in liver, although the mRNA expression was opposite. As shown in Fig. 6, there was interindividual variability in AADAC protein level and flutamide hydrolase activity in human liver. The discrepancy may be because of use of liver RNA derived from single donor. In general, the small intestine plays an important role in the first-pass metabolism of therapeutic drugs given orally (Lin et al., 1999). AADAC would play a certain role in the first-pass metabolism of flutamide in small intestine and in liver.
Correlation analysis was performed between the expression level of AADAC protein and the flutamide hydrolase activity using individual HLM samples (Fig. 6). The correlation between the expression level of AADAC and the flutamide hydrolase activity was strongly significant. Although the point appears to show an x-axis intercept greater than 0, several individual HLM samples may partially include inactive AADAC protein. The expression level of AADAC protein and the flutamide hydrolase activity were moderately variable. It is feasible that the induction by xenobiotics from the environment or diet and endobiotics, and genetic polymorphism of the AADAC gene affect the interindividual variability of the flutamide hydrolase activity and AADAC expression level. However, the regulation mechanisms of human AADAC expression are not fully understood. Saito et al. (2003) previously found 23 SNPs in the AADAC gene using DNA samples of 48 Japanese. Among them, only an SNP that was also found in this study (g.13651G > A, c.931G > A) leads to an amino acid change (V281I). However, AADAC variant (V281I) appeared not to alter the enzyme activity (Fig. 3B). It is considered that flutamide hydrolysis is important in the occurrence of hepatotoxicity (Aizawa et al., 2003; Matsuzaki et al., 2006; Ohbuchi et al., 2009). Therefore, the interindividual variability of AADAC might affect the incidence of flutamide-induced hepatotoxicity. Further study on the regulation mechanisms and genetic polymorphisms of human AADAC will be necessary.
In conclusion, we found that human AADAC is a principal enzyme in the flutamide hydrolysis. The present study is the first report of the contribution of human AADAC to the metabolism of a therapeutic drug.
Acknowledgments
We thank Brent Bell for review of the manuscript.
Footnotes
-
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
-
doi:10.1124/dmd.109.026567.
-
ABBREVIATIONS: FLU-1, 4-nitro-3-(trifluoromethyl)phenylamine; CES, carboxylesterase; HLM, human liver microsome; BNPP, bis-(nonylphenyl)-phenylphosphate; AADAC, arylacetamide deacetylase; DFP, diisopropylphosphorofluoride; eserine, physostigmine sulfate; PMSF, phenylmethylsulfonyl fluoride; AgNO3, silver nitrate; CdCl2, cadmium chloride; CoCl2, cobaltous chloride; CuCl2, cupric chloride; PNPA, p-nitrophenyl acetate; NaF, sodium fluoride; HLC, human liver cytosol; HJM, human jejunum microsome; HPM, human pulmonary microsome; HRM, human renal microsome; RT-PCR, reverse transcription-polymerase chain reaction; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; SNP, single nucleotide polymorphism; DMSO, dimethyl sulfoxide; HPLC, high-performance liquid chromatography.
- Accepted March 26, 2009.
- Received January 19, 2009.
- The American Society for Pharmacology and Experimental Therapeutics