Abstract
The initial glucuronidation rates were determined for eight recombinant human UDP-glucuronosyltransferases (UGTs) of the 1A subfamily, and the bisubstrate kinetics and inhibition patterns were analyzed. At low substrate concentrations, the reactions followed general ternary complex kinetics, whereas at higher concentrations of both substrates, the reactions were mostly characterized by ternary complex kinetics with substrate inhibition. The glucuronidation of entacapone by UGT1A9 was inhibited by 1-naphthol in a competitive fashion, with respect to entacapone, and an uncompetitive fashion, with respect to UDP-glucuronic acid (UDPGA). Its inhibition by UDP, on the other hand, was noncompetitive with respect to entacapone and competitive with respect to UDPGA. These inhibition patterns are compatible with a compulsory ordered bi bi mechanism in which UDPGA is the first-binding substrate. Despite the identical primary structure of the C-terminal halves of the UGT1A isoforms, there were marked differences in the respective Km values for UDPGA, ranging from 52 μM for UGT1A6 to 1256 μM for UGT1A8. Relative specificity constants were calculated for the eight UGT1A isoforms with 1-hydroxypyrene, 4-nitrophenol, scopoletin, 4-methylumbelliferone, and entacapone as aglycone substrates. The results demonstrated that seven of the UGT1A isoforms are capable of conjugating phenolic substrates with similar highest kcat values, and UGT1A4 has a lower relative turnover rate. The highest specificity constants were obtained for 1-hydroxypyrene, even with UGT1A6, which has been regarded as a specific isoform for small planar phenols. A kcat value of 1.9 s–1 was calculated for the glucuronidation of scopoletin by purified UGT1A9.
UDP-glucuronosyltransferases (UGTs; EC 2.4.1.17) catalyze the transfer of glucuronic acid from the cosubstrate uridine 5′-diphospho-α-d-glucuronic acid (UDPGA) to various aglycone compounds, such as steroid hormones, bile acids, and bilirubin, as well as to a large number of xenobiotics including drugs and drug metabolites.
Expressed in the liver and several extrahepatic tissues (Tukey and Strassburg, 2000), human UGTs are integral membrane proteins that reside within the endoplasmic reticulum, with their catalytic sites, as well as most of their mass, on the lumenal side of the membrane. They have a single trans-membrane segment close to their C terminus and their last 19 to 26 residues are located on the cytoplasmic side of the endoplasmic reticulum membrane. UGTs appear to be built of two large domains of almost equal size, namely, the N- and C-terminal domains. At least three subfamilies, UGT1A, UGT2A, and UGT2B, are distinguished on the basis of sequence homology and gene structure (Mackenzie et al., 1997). The 1A subfamily comprises nine functional isoforms. The N-terminal domains of UGT1A isoforms are encoded by different first exons and their primary structures are more variable. The C-terminal domains are encoded by the same exons 2 to 5, so that the primary structure of the C-terminal domains, roughly corresponding to the C-terminal halves, are identical in all UGT1A isoforms (Gong et al., 2001).
This very high degree of homology of the C-terminal halves of all the UGTs, as well as interactions of periodate-oxidized UDPGA with UGT1A6 (Battaglia et al., 1998), strongly suggests that the C-terminal domain is directly involved in UDPGA binding, whereas the N-terminal halves contain the aglycone binding site(s) (Mackenzie, 1990; Senay et al., 1999; Gong et al., 2001). It has been suggested, however, that the N-terminal part of the mature protein may also participate in UDPGA binding (Pillot et al., 1993). In addition, recent findings suggest that protein-protein interactions between the N-terminal and C-terminal halves of the UGTs may have functional importance, perhaps providing a mechanism for one of these two major domains to affect substrate or cosubstrate binding in the other (Kurkela et al., 2004).
The most common glucuronidation sites are nucleophilic oxygen or nitrogen atoms of the sugar acceptor substrate. Glucuronidation occurs as an SN2 substitution by the attack of a nucleophilic heteroatom of the aglycone on the C1 atom of glucuronic acid, whereas both UDPGA and the aglycone substrate are bound on the active site of the enzyme. The SN2 mechanism is supported by inversion of the α-configuration of the C1 atom to β-configuration in the glucuronide (Johnson and Fenselau, 1978), and by the substituent effects on the glucuronidation rate (Yin et al., 1994). The three-dimensional structure of the active site is unknown. A unanimous conclusion of all previous mechanistic studies is that the reaction involves the formation of a ternary complex (Potrepka and Spratt, 1972; Vessey and Zakim, 1972; Sanchez and Tephly, 1975; Rao et al., 1976; Koster and Noordhoek, 1983; Falany et al., 1987; Matern et al., 1991; Yin et al., 1994). Product and dead-end inhibition studies conducted to determine the order of substrate binding have given ambiguous results, however. Two reports based on partially purified rat UGTs (Falany et al., 1987; Yin et al., 1994), and one report based on beef liver microsomes (Vessey and Zakim, 1972), have proposed a random ordered bi bi mechanism, whereas other studies with animal tissue fractions have proposed a compulsory ordered bi bi (Potrepka and Spratt, 1972; Sanchez and Tephly, 1975; Koster and Noordhoek, 1983) or a Theorell-Chance mechanism (Rao et al., 1976; Koster and Noordhoek, 1983).
The availability of recombinant UGTs has aided in understanding the contribution of glucuronidation to xenobiotic metabolism. Substrate specificities of human UGTs have been characterized by determining specific activities and kinetic parameters for a large number of substrates. Nevertheless, the role and significance of individual UGT isoforms are still poorly understood, and the bisubstrate kinetics of UGTs has not previously been studied with individual isoenzymes. Meaningful substrate specificity assessment can only be made after the catalytic constants (kcat), and especially the specificity constants (kcat/Km), have been determined, or at least normalized values have been assigned for individual isoenzymes. The other parameters of the bisubstrate kinetics should also be determined. Detailed study of the kinetic mechanism of UGTs could also assist the design of potent and specific inhibitors.
To better understand the kinetics of glucuronidation, we studied the bisubstrate kinetics of eight human recombinant UGTs of the 1A subfamily. The kinetic mechanism and substrate inhibition were characterized and the substrate specificities were evaluated on the basis of relative specificity constants.
Materials and Methods
Materials. Insect cell membranes containing one of the following human UGTs, 1A1, 1A3, 1A4, and 1A6–1A10, together with purified UG1A9, were prepared in our laboratory as previously described (Kurkela et al., 2003; Kuuranne et al., 2003). Phosphatidylcholine X-E from dried egg yolk, ethinylestradiol, scopoletin, 4-aminobiphenyl, 1-naphthol, 4-hydroxyestrone, 4-nitrophenol, umbelliferone, uridine 5′-diphosphate sodium salt (UDP), d-saccharic acid 1,4-lactone (saccharolactone), and uridine 5′-diphosphoglucuronic acid trisodium salt (UDPGA) were purchased from Sigma-Aldrich (St. Louis, MO), and 1-hydroxypyrene was obtained from Acros Organics (Fairlawn, NJ). Entacapone was kindly provided by Orion Pharma (Espoo, Finland), and entacapone glucuronide (Luukkanen et al., 1999), 1-hydroxypyrene glucuronide (Luukkanen et al., 2001), and scopoletin glucuronide were synthesized in our laboratory. α-Naphthyl-β-d-glucuronide, 4-nitrophenyl-β-d-glucuronide, and umbelliferyl-β-d-glucuronide were purchased from Sigma-Aldrich. Radiolabeled [14C]UDPGA was obtained from PerkinElmer Life and Analytical Sciences (Boston, MA).
Activity Assays. All enzyme assays (total volume 250 μl) contained 50 mM phosphate buffer, pH 7.4, 5 mM MgCl2, 5 mM saccharolactone, 25 to 5000 μM UDPGA, and 10 to 50 μg of membrane protein of the appropriate recombinant UGT or 50 ng of purified UGT1A9. Phosphatidylcholine type X-E (1 mg/ml) was added to the assay mixtures containing the purified UGT1A9. The aglycone substrate concentrations ranged from ca. 0.2 to 10 times the respective Km values (determined in preliminary assays) unless method sensitivity or substrate solubility demanded otherwise. The aglycone substrates were added as DMSO solutions to a final DMSO concentration of 2% in all the enzyme assays. All samples were prepared in duplicate.
The reactions were performed at 37°C and the reaction time varied, within the linear range, from 10 to 60 min. The reactions were terminated by the addition of 25 μl of 4 M ice-cold perchloric acid or 250 μl of ice-cold methanol (4-aminobiphenyl). The precipitated proteins were removed by centrifugation (5 min at 14,000 rpm). Blank incubations were prepared in a similar way at the highest aglycone substrate concentrations without UDPGA.
Aliquots of the supernatants were analyzed using an Agilent model 1100 (Agilent Technologies, Palo Alto, CA) or a Hewlett Packard model 1090 HPLC apparatus (Hewlett Packard, Palo Alto, CA) with UV or fluorescence detection. Details of the analytical conditions are given in Table 1. The quantitation of entacapone glucuronide, 1-naphthylglucuronide, 1-hydroxypyrene glucuronide, scopoletin glucuronide, and 4-nitrophenylglucuronide was based on authentic reference standards. When pure glucuronide was not available, the quantitation of the glucuronidated products was achieved with the HPLC apparatus connected to a flow scintillation analyzer (150TR; PerkinElmer Life and Analytical Sciences) that was fitted with a 500-μl flow cell into which 3 ml/min scintillation liquid (Monoflow 3; National Diagnostics, Atlanta, GA) was pumped. The radiochemical detection was coupled to UV or fluorescence detection as described previously (Kaivosaari et al., 2001).
Analytical conditions in the kinetic analysis of eight UGT1A isoenzymes
Product inhibition studies with UDP (UGT1A9) were conducted at subsaturating (100 μM) concentrations of the fixed substrate. Inhibition studies with a competing substrate, 1-naphthol, were conducted at 100 μM entacapone when UDPGA was the varied substrate, and at 1000 μM UDPGA when entacapone was the varied substrate. UDP was added to the reaction mixture as an aqueous solution to a final concentration of 0, 25, 50, 100, 250, or 500 μM, and 1-naphthol in DMSO was added to a final concentration of 0, 0.5, 2.5, or 10 μM.
Protein concentrations were measured using the BCA protein assay kit (Pierce Chemical, Rockford, IL) with bovine serum albumin as standard, and the relative enzyme concentrations in the membranes were estimated by dot-blot analyses as described recently (Kurkela et al., 2004).
Kinetic Analysis. Kinetic parameters were estimated by fitting the initial rate data to appropriate bisubstrate and inhibition rate equations (Cornish-Bowden, 1995; Copeland, 2000) by nonlinear least-squares regression using a weighting factor 1/y. Data analysis was performed by SigmaPlot Enzyme kinetics module 1.1S (SPSS Inc., Chicago, IL), and the goodness of fit was evaluated on the basis of standard deviations of the parameter estimates, R2 values, and residual plots. The individual rate constants for the glucuronidation of scopoletin by purified UGT1A9 were calculated according to the method of Cornish-Bowden (1995).
Results
Kinetic Mechanism. The initial rates for the reactions catalyzed by the recombinant human UGTs 1A1, 1A3, 1A4, 1A6, 1A7, 1A8, 1A9, and 1A10 were determined by varying the concentrations of both substrates: the sugar donor UDP-glucuronic acid (AX) and the sugar acceptor (B). The measured initial rates were in good agreement with a ternary complex mechanism, for which the reaction velocity can be given by eq. 1 or eq. 2 (Copeland, 2000), where KAX is the dissociation constant for the enzyme-AX complex, KB is the dissociation constant for the enzyme-B complex, and KmAX and KmB are the Michaelis constants for AX and B, respectively. 
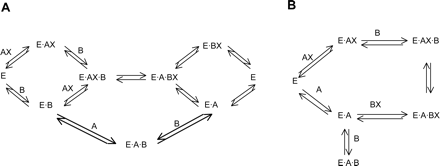
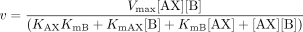 Equation 1 is derived for the random ordered bi bi mechanism (see Fig. 2A), assuming rapid equilibrium, and eq. 2 is derived for the compulsory ordered bi bi mechanism (see Fig. 2B), using steady-state assumptions.
Equation 1 is derived for the random ordered bi bi mechanism (see Fig. 2A), assuming rapid equilibrium, and eq. 2 is derived for the compulsory ordered bi bi mechanism (see Fig. 2B), using steady-state assumptions.
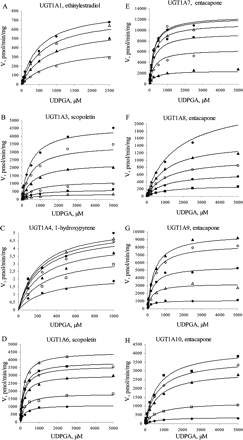
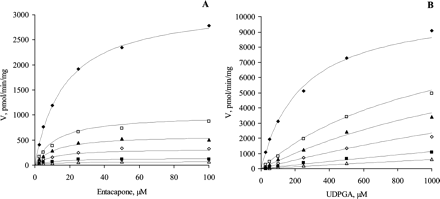
The bisubstrate initial rate data for all UGT1A enzymes gave a good statistical fit to both eqs. 1 and 2 up to aglycone substrate concentrations at least 4 to 5 times the respective Km values (Table 2) and up to UDPGA concentrations of 1 to 5 mM. Fit of the kinetic data to eq. 2 is shown in Fig. 1. At higher aglycone concentrations, substrate inhibition was observed for all UGT1A isoforms except UGT1A4. At very high concentrations of both substrates (above 2 mM), the measured velocity data showed irregular behavior and poor reproducibility. In the case of UGT1A3, the high Km for scopoletin limited the rate determination to a concentration of aglycone of only twice the Km.
The enzyme kinetic parameters for eight UGT1A isoenzymes with appropriate aglycone substrates
The initial rate data were fitted to eq. 2 omitting the high substrate concentrations with obvious substrate inhibition.
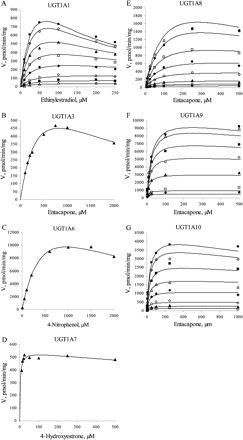
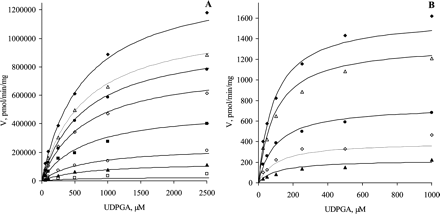
Substrate inhibition is expected for a ternary complex mechanism following compulsory ordered kinetics. As depicted in Fig. 2B, binding of the aglycone substrate to the enzyme-UDP complex leads to a nonproductive dead-end complex that slows the completion of the catalytic cycle. In such a case, the reaction velocity is given by eq. 3: 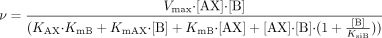 where KsiB is a constant that describes the strength of substrate inhibition. The bisubstrate kinetic data for 1A1, 1A8, 1A9, and 1A10, including conditions of high aglycone substrate concentration, fitted well to eq. 3 (Fig. 3). In the case of UGTs 1A3, 1A6, and 1A7, the substrate inhibition data were measured only at one UDPGA concentration, and the data fitted well to the corresponding reduced equation (eq. 4).
where KsiB is a constant that describes the strength of substrate inhibition. The bisubstrate kinetic data for 1A1, 1A8, 1A9, and 1A10, including conditions of high aglycone substrate concentration, fitted well to eq. 3 (Fig. 3). In the case of UGTs 1A3, 1A6, and 1A7, the substrate inhibition data were measured only at one UDPGA concentration, and the data fitted well to the corresponding reduced equation (eq. 4). 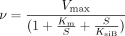 Although formation of the same nonproductive complex is also possible in a mechanism that follows random ordered ternary complex kinetics (Fig. 2A), the formation should not occur and substrate inhibition should not be seen if the mechanism involves rapid equilibrium, because then, the concentration of the enzyme-UDP or enzyme-glucuronide complex is zero in the absence of added products. Observation of substrate inhibition may thus be used to exclude the rapid equilibrium random ordered bi bi mechanism.
Although formation of the same nonproductive complex is also possible in a mechanism that follows random ordered ternary complex kinetics (Fig. 2A), the formation should not occur and substrate inhibition should not be seen if the mechanism involves rapid equilibrium, because then, the concentration of the enzyme-UDP or enzyme-glucuronide complex is zero in the absence of added products. Observation of substrate inhibition may thus be used to exclude the rapid equilibrium random ordered bi bi mechanism.
The results of the bisubstrate kinetic experiments suggest that glucuronidation catalyzed by UGTs 1A1, 1A3, and 1A6-1A10 does not involve rapid equilibrium, and the reaction most probably follows the scheme presented in Fig. 2B. Nevertheless, a non-rapid-equilibrium random ordered reaction according to Fig. 2A cannot be excluded on the basis of the bisubstrate kinetic data alone.
Two other possible mechanisms, rapid equilibrium compulsory ordered mechanism (Theorell-Chance) and the double displacement mechanism (Ping-Pong) (Copeland, 2000) can be excluded on the basis of the poor fit of the velocity data to the corresponding equations.
An inhibitor that binds to the same site as one of the substrates would kinetically distinguish between the two ternary complex mechanisms. In the case of the compulsory ordered mechanism, an inhibitor that resembles the second-binding substrate would be expected to bind to the enzyme–first-binding substrate complex rather than to the free enzyme. Hence, a competitive inhibitor with respect to the second-binding substrate would behave as an uncompetitive inhibitor with respect to the first-binding substrate. In the case of a random ordered mechanism, on the other hand, no uncompetitive inhibition pattern should be observed. Carrying of such an inhibition experiment has been hampered by the lack of effective UGT inhibitors that bind to the aglycone-binding site. However, we found that 1-naphthol, although it is a substrate for UGT1A9, shows significantly higher affinity and lower reactivity (Km = 0.09 μM, Vmax = 27.8 pmol/ml/min) than does entacapone (Table 2) and, hence, could be used as a competitive inhibitor with respect to entacapone in experiments with UGT1A9.
The effects of 1-naphthol on the kinetics of entacapone glucuronidation by UGT1A9 are shown in Fig. 4. As can be seen, 1-naphthol acts as an uncompetitive inhibitor when UDPGA is the varied substrate (Fig. 4A), and as a competitive inhibitor when entacapone is the varied substrate (Fig. 4B). Fitting the data to the general equation of reversible inhibition yielded dissociation constants of 0.191 μM and 1.46 μM for the binding of 1-naphthol to the enzyme-UDPGA complex and to the free enzyme, respectively. These results indicate that 1-naphthol also binds to the free enzyme, but with significantly lower affinity than to the enzyme-UDPGA complex. It is not unexpected to see some inhibitory binding of a hydrophobic substrate to the free enzyme, even though the catalytic binding would require compulsory order.
We also studied UDP inhibition of the entacapone glucuronidation reaction by UGT1A9. The results are presented in Fig. 5. The inhibition by UDP displayed noncompetitive and competitive patterns with respect to entacapone (Fig. 5A) and UDPGA (Fig. 5B), respectively. Such inhibition patterns are possible in both random and compulsory ordered mechanisms. In the case of a compulsory ordered reaction, the inhibitory behavior of UDP suggests that UDPGA is the first-binding substrate. The dissociation constant Ki for enzyme-UDP complex was found to be less than one-tenth the Ki value for the enzyme-UDPGA complex (Fig. 5).
Kinetic Parameters. The enzyme kinetic parameters for the glucuronidation of appropriate aglycone substrates by the eight UGT1A isoforms were determined using eq. 2. The results are listed in Table 2. The initial velocities obtained at high substrate concentrations with obvious substrate inhibition were omitted from this kinetic analysis. Considerable variation was observed in UDPGA parameter values for the different UGT1A isoforms. The lowest Km value, 52 μM, was determined for the glucuronidation of 1-naphthol by UGT1A6 and the highest, 1256 μM, for the glucuronidation of 4-hydroxyestrone by UGT1A8. The KAX values, the dissociation constants of the enzyme-UDPGA complex, in these cases were 163 μM and 1264 μM, respectively. In the compulsory ordered mechanism, the KAX value should be independent of the aglycone substrate, if UDPGA is the first-binding substrate. To test this hypothesis, the kinetic parameters for UGT1A6 and UGT1A8 were also determined for the glucuronidation of scopoletin (166 μM) and entacapone (875 μM), respectively, and the results are included in Table 2. The KAX values for the latter substrates are similar to those obtained with 1-naphthol (1A6) and 4-hydroxyestrone (1A8), as expected according to the proposed mechanism.
The bisubstrate initial rate data showing obvious substrate inhibition were fitted to eq. 3 and, when only the aglycone substrate was varied, to the corresponding reduced equation. The kinetic parameters Km, Vmax, KAX, and KsiB are presented in Table 3. The Km, Vmax, and KAX values are mostly in reasonable agreement with the values given in Table 2. The KsiB values varied from 178 μM for the glucuronidation of ethinylestradiol by UGT1A1 to 2800 μM for the glucuronidation of entacapone by UGT1A10. In all these cases the KsiB values were significantly higher than the corresponding Km values.
The enzyme kinetic parameters for seven UGT1A isoforms showing substrate inhibition
The initial rate data were fitted to eq. 3 or 4.
Relativekcat and Relative Specificity Constants. The relative specificity constants allow a comparison of the catalytic efficiencies of different enzymes or of the utilization of different substrates by the same enzyme. The relative kcat and relative kcat/Km values for 1-hydroxypyrene, 4-nitrophenol, scopoletin, 4-methylumbelliferone, and entacapone with eight UGT1A isoforms are presented in Table 4. Relative kcat values and relative specificity constants, kcat/Km, were calculated on the basis of the relative expression levels of the individual UGT1A isoforms. The kcat and kcat/Km values for the glucuronidation of entacapone by UGT1A9 were arbitrarily assigned as 1.0, and the values for other UGTs were compared with those for UGT1A9. In general, all UGT1A isoforms were capable of conjugating phenolic substrates, and the highest relative kcat values for all isoforms, with the exception of 1A4, were closely similar. UGT1A4 also conjugated phenolic substrates but at lower efficiency. Nevertheless, examination of the relative kcat/Km values revealed that, regardless of the relative kcat values per se, all eight UGTs showed the highest relative kcat/Km values with 1-hydroxypyrene, a large polycyclic aromatic planar phenol.
Relative catalytic efficiency of eight recombinant human UGTs of the 1A subfamily in glucuronidation of phenolic compounds
Relative catalytic constant (kcat = 1.0) and relative specificity constant (kcat/Km = 1.0) were arbitrarily assigned for the glucuronidation of entacapone by UGT1A9.
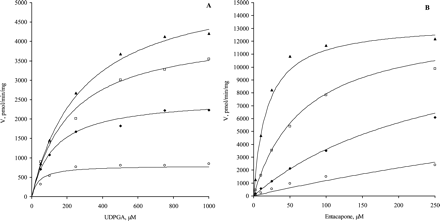
Catalytic Constantkcat. A kcat value of 1.9 s–1 was estimated from kinetic experiments with purified UGT1A9 (Fig. 6). A consistent means of determining kcat requires knowledge of the absolute enzyme concentration and, hence, the total UGT1A9 concentration was calculated by means of the total amount of protein in the purified enzyme. The apparent homogeneity of the purified UGT1A9 was demonstrated by SDS-polyacrylamide gel electrophoresis and Western blotting (Kurkela et al., 2003).
In the case of the compulsory ordered bi bi mechanism, a unique relationship exists between the kinetic parameters and the rate constants (Fig. 7). Hence, the individual rate constants k1 and k–1 could be calculated from the initial rate measurements (Cornish-Bowden, 1995).
The rate constants for the glucuronidation of scopoletin by purified UGT1A9 are presented in Fig. 6. The first order rate constant for the dissociation of the enzyme-UDPGA complex, k–1 = 2.1 s–1, equals the rate constant for the product formation, kcat, in accordance with the steady-state assumption.
The bisubstrate kinetics for eight human recombinant UGTs. The initial rate data were fitted to the rate equation for ternary complex mechanism (eq. 2) at concentration ranges with negligible substrate inhibition; the R2 values varied from 0.959 to 0.998. A, UGT1A1 (10, 25, 50, and 100 μM ethinylestradiol); B, UGT1A3 (25, 50, 100, 250, 500, and 1000 μM scopoletin); C, UGT1A4 (10, 25, 50, 100, 150, and 250 μM 1-hydroxypyrene); D, UGT1A6 (50, 100, 250, 400, 500, and 1000 μM scopoletin); E, UGT1A7 (5, 20, 50, 100, 200, and 250 μM entacapone); F, UGT1A8 (10, 25, 50, 100, and 250 μM entacapone); G, UGT1A9 (2.5, 10, 50, 100, and 250 μM entacapone); H, UGT1A10 (2.5, 10, 50, 100, and 250 μM entacapone). Each data point represents the average of at least two replicates. Experimental details are presented under Materials and Methods. For kinetic parameters, see Table 2.
Scheme A presents a random ordered bi bi mechanism and scheme B a compulsory ordered bi bi mechanism. E = UGT, AX = UDP-glucuronic acid, B = aglycone substrate.
Discussion
The bisubstrate kinetics of eight recombinant human UGTs was analyzed. The glucuronidation reaction was well described by the rate equations derived for mechanisms involving the formation of a ternary complex. Nevertheless, the bisubstrate kinetics alone did not allow us to determine whether there was a compulsory order of binding of substrates. An unambiguous differentiation of the random and compulsory ordered mechanisms was established by the observed, essentially uncompetitive inhibition by 1-naphthol with respect to UDPGA (Fig. 4A). This result indicates that 1-naphthol preferably binds to the enzyme-UDPGA complex; that is, the reaction follows a compulsory ordered kinetic mechanism in which UDPGA is the first-binding substrate. The observed competitive inhibition by UDP with respect to UDPGA (Fig. 5B) and noncompetitive inhibition with respect to entacapone (Fig. 5A) provided further evidence for the compulsory ordered mechanism with UDPGA as the first-binding substrate.
Previous studies on the enzyme kinetic mechanism have provided different and sometimes conflicting results (Potrepka and Spratt, 1972; Vessey and Zakim, 1972; Sanchez and Tephly, 1975; Rao et al., 1976; Koster and Noordhoek, 1983; Falany et al., 1987; Yin et al., 1994). The ambiguity of the results from product and dead-end inhibition experiments in earlier work may well be explained by the presence of several different UGT isoforms in the tissue samples, a fact that was not fully appreciated in the past. The presence of several isoforms would severely complicate the interpretation of inhibition kinetic data, especially when analyzed by means of linearized plots. In addition, those studies that relied on partially purified rat enzymes probably suffered from the presence of UGTs that were partially detergent-inactivated during the membrane solubilization and the purification process (Kurkela et al., 2003).
At high concentrations of both substrates, substrate inhibition was frequently observed with all UGT1A isoforms except 1A4. The only human UGT of the 1A subfamily for which we have not detected substrate inhibition was 1A4. This enzyme preferably catalyzes the N-glucuronidation of aglycones containing amino groups, and only a few phenolic compounds are glucuronidated by UGT1A4 efficiently. An attempt was made to determine the enzyme kinetic parameters for the glucuronidation of 4-aminobiphenyl, which is a widely accepted model substrate for UGT1A4. Although it was a good substrate for UGT1A4 when considering Vmax, the bisubstrate kinetics for 4-aminobiphenyl could not be determined reliably due to the combination of low solubility and poor method sensitivity. A hydroxylated polycyclic aromatic hydrocarbon compound, 1-hydroxypyrene, exhibited a considerably lower Km but showed no substrate inhibition in the concentration range that was studied. The apparent substrate inhibition was used to exclude the rapid equilibrium random ordered bi bi mechanism. In the case of the compulsory ordered bi bi mechanism, substrate inhibition allows the identification of the second-binding substrate. The occurrence of substrate inhibition for UGTs 1A1, 1A3, and 1A6-1A10 suggests, therefore, that glucuronidation catalyzed by these isoforms does not involve rapid equilibrium, and that the reaction probably follows the scheme presented in Fig. 2B. The KsiB values (Table 3) were significantly higher than the corresponding Km values for all UGT1A isoforms, however, which means that substrate inhibition is probably of negligible significance in a physiological environment. As reported previously (Houston and Kenworthy, 2000; Uchaipichat et al., 2004), substrate inhibition may also arise from the binding of a second substrate molecule to the enzyme-substrate complex. Nevertheless, the kinetic data fitted well to eqs. 3 and 4, and incorporation in the kinetic equation of a parameter describing the binding of a second substrate molecule resulted in poorer statistics of the fits.
Kinetic constants derived from bisubstrate reactions have not been reported previously for individually expressed UGT isoforms. The Km values for UDPGA have been determined at saturating aglycone substrate concentrations with recombinant human UGTs 1A1 (400–1810 μM) (Senafi et al., 1994; Court et al., 2001), 1A3 (250 μM), 1A4 (300 μM) (Green et al., 1995), 1A6 (110–200 μM) (Battaglia et al., 1998; Ouzzine et al., 2000; Court et al., 2001), and 1A9 (190–500 μM) (Senafi et al., 1994; Court et al., 2001). The Km values obtained in this work (Table 2) are within the same range as most of those results. The physiological concentration of UDPGA in human liver is ca. 300 μM (301 μmol/kg wet weight) and at least 1 order of magnitude lower than this in human kidney (Cappiello et al., 2000). The physiological concentrations of UDPGA only rarely exceed the Km values obtained in this study, and speculations that a decrease in the availability of UDPGA would impair the capacity for hepatic glucuronide conjugation (Howell et al., 1986; Evdokimova et al., 2001) may be justified. A comparison of the aglycone Km values in Table 2 with the few published results for the same combinations of UGT isoform and substrate shows our values to be mostly within the same order of magnitude as the earlier ones, for example, those reported for UGT1A1 with ethinylestradiol (Williams et al., 2002; Soars et al., 2003), UGT1A3 with scopoletin (Green et al., 1998), and UGT1A6 with 1-naphthol (Uchaipichat et al., 2004), as well as those reported for eight UGT1A isoforms with 4-methylumbelliferone (Uchaipichat et al., 2004).
The substrate inhibition kinetics for seven human recombinant UGTs. The initial rate data were fitted to the rate equation for the ternary complex mechanism, taking into account substrate inhibition (eqs. 3 and 4). Initial rate data were either from the bisubstrate kinetic analysis (25, 50, 100, 250, 500, 1000, 2500, and 5000 μM UDPGA) or determined at 1 mM UDPGA. The R2 values varied from 0.864 to 0.994. A, UGT1A1 (ethinylestradiol); B, UGT1A3 (entacapone); C, UGT1A6 (4-nitrophenol); D, UGT1A7 (4-hydroxyestrone); E, UGT1A8 (entacapone); F, UGT1A9 (entacapone); G, UGT1A10 (entacapone). Each data point represents the average of at least two replicates. Experimental details are presented under Materials and Methods. For kinetic parameters, see Table 3.
Effect of 1-naphthol on entacapone glucuronidation. Inhibition by a competing substrate was determined for UGT1A9 at 100 μM entacapone when UDPGA was the varied substrate (0, 0.05, 0.2, and 1 μM 1-naphthol) and at 1000 μM UDPGA when entacapone was the varied substrate (0, 0.5, 2.5, and 10 μM 1-naphthol). 1-Naphthol was an uncompetitive inhibitor (A) with respect to UDPGA (Ki = 0.169 ± 0.004 μM), and a competitive inhibitor (B) with respect to entacapone (Ki = 0.183 ± 0.0033 μM). Each data point represents the average of two replicates.
Inhibition of entacapone glucuronidation by UDP. Product inhibition by UDP (0, 25, 50, 100, 250, and 500 μM) was determined for UGT1A9 at a 100 μM concentration of the fixed substrate. UDP was a noncompetitive inhibitor (A) with respect to entacapone (Ki = 20.3 ± 1.03 μM, αKi = 11.5 μM) and competitive (B) with respect to UDPGA (Ki = 6.03 ± 0.135 μM). Each data point represents the average of two replicates.
The current study also has implications for the structure-function relationships of the UGTs. A close inspection of the KAX values in Table 2 reveals distribution to low and high values. UGTs 1A4, 1A6, 1A7, and 1A9 exhibited low KAX values (163–316 μM), whereas UGTs 1A1, 1A3, 1A8, and 1A10 exhibited higher KAX values (693–1264 μM) for UDPGA. A similar distribution of UGTs appears in the Km values, with the exception of UGT1A1, which had low Km and higher KAX (Table 2). These differences in KAX values suggest that the three-dimensional structure of the UDPGA binding site is not fully conserved, not even within the UGT1A subfamily. Furthermore, UGTs 1A7-1A10 are highly homologous throughout the entire protein, and this is reflected in similar Km values toward entacapone. Nevertheless, UGT1A9 has very low KAX and low Km for UDPGA, whereas UGT1A8 exhibits the highest KAX and Km values in this study (Table 2). These results suggest that, in addition to the C-terminal half of the UGTs, the UDPGA binding is affected by the amino acid residues of the N-terminal half of the protein.
The relative kcat values (Table 4) demonstrate that all the UGT1A isoforms are capable of conjugating phenolic substrates, and all isoforms with the exception of UGT1A4 exhibit similar kcat values for their best phenolic substrates. All the UGT1A isoforms were efficient catalysts of 1-hydroxypyrene and scopoletin glucuronidation, but UGT1A6 was the only isoform that could conjugate 4-nitrophenol with high efficiency. Entacapone, with its large nonplanar side chain, was a poor substrate for UGTs 1A1-1A6 and a good substrate for UGTs 1A7-1A10. The highest relative specificity constants with all isoforms were observed for the glucuronidation of 1-hydroxypyrene, a large polycyclic aromatic planar phenol. These results suggest, contrary to the general assumption, that UGT1A6 is not restricted to the glucuronidation of small planar phenols.
Kinetics of purified and membrane-bound UGT1A9. A, bisubstrate kinetics of purified UGT1A9 at 10, 50, 100, 250, 500, 750, 1000, and 2000 μM scopoletin as aglycone substrate. The kinetic parameters were: Km (UDPGA) = 623 ± 2.37 μM, Km (scopoletin) = 673 ± 2.37 μM, Vmax = 1885 ± 3.33 nmol/min/mg, KiA = 689 ± 2.68 μM, KiB = 745 ± 3.15 μM, kcat = 1.9 s–1, k1 = 3026 M–1 s–1, k–1 = 2.1 s–1. All data points represent a single determination. B, bisubstrate kinetics of membrane-bound UGT1A9 at 5, 10, 25, 100, and 250 μM scopoletin as aglycone substrate. See Table 2 for kinetic parameters. All data points are the average of two replicates.
A scheme presenting a compulsory ordered bi bi mechanism and individual rate constants. AX, UDPGA; B, aglycone substrate; A, UDP; BX, glucuronide; kcat = k3k4/(k3 + k4).
The individual rate constants for the binding of UDPGA to the enzyme, the dissociation of the enzyme-UDPGA complex, and the product formation for the glucuronidation of scopoletin by purified UGT1A9 were calculated from the enzyme kinetic parameters (Fig. 6A), assuming the enzyme follows the compulsory ordered mechanism. The purified UGT1A9 presented a lower affinity for scopoletin and UDPGA than the native protein, as evidenced by significant increases in the Km and KAX values. Nevertheless, the specific activity of the purified enzyme was ca. 1000-fold higher than the value of the membrane-bound UGT1A9.
When [S]≪Km, the upper limit for the reaction rate is defined by the diffusional rate of encounter of the free enzyme with substrate, which is characterized by k1 (typically 108 to 109 M–1 s–1). When [S]≫Km, in turn, the formation of [ES] complex is rapid and often not rate-limiting. Here, the formation of the enzyme-UDPGA complex was rapid (3026 M–1 s–1) compared to the formation of scopoletin glucuronide (1.9 s–1) (Fig. 6). The kcat value (1.9 s–1) for the formation of scopoletin glucuronide was 1 to 2 orders of magnitude higher than previously reported for the N-glucuronidation of N-hydroxy-PhIP by UGT1A9 (Malfatti and Felton, 2004). However, UGT1A1 catalyzed the glucuronidation of N-hydroxy-PhIP at the highest rate, kcat = 1.9 s–1, which is in line with our observation that UGTs 1A1, 1A3, and 1A6-1A10 show similar kcat values for their best phenolic substrates.
Acknowledgments
We thank Sanna Sistonen for skillful technical assistance and Orion Pharma for generously providing entacapone.
Footnotes
-
This work was supported by the Technology Development Center of Finland (TEKES) and the Academy of Finland (project 207535).
-
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
-
doi:10.1124/dmd.105.004093.
-
ABBREVIATIONS: UGT, UDP-glucuronosyltransferase; UDPGA, UDP-glucuronic acid; DMSO, dimethyl sulfoxide; HPLC, high-performance liquid chromatography; N-hydroxy-PhIP, N-hydroxy-2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine.
- Received February 8, 2005.
- Accepted March 30, 2005.
- The American Society for Pharmacology and Experimental Therapeutics