Abstract
The specific cytochrome P450 (P450) isoforms mediating the biotransformations of clobazam (CLB) and those of its major metabolites, N-desmethylclobazam (NCLB) and 4′-hydroxyclobazam were identified using cDNA-expressed P450 and P450-specific chemical inhibitors. Among the 13 cDNA-expressed P450 isoforms tested, CLB was mainly demethylated by CYP3A4, CYP2C19, and CYP2B6 and 4′-hydroxylated by CYP2C19 and CYP2C18. CYP2C19 and CYP2C18 catalyzed the 4′-hydroxylation of NCLB. The kinetics of the major biotransformations were studied: CYP3A4, CYP2C19, and CYP2B6 mediated the formation of NCLB with Km = 29.0, 31.9, and 289 μM, Vmax = 6.20, 1.15, and 5.70 nmol/min/nmol P450, and intrinsic clearance (CLint) = 214, 36.1, and 19.7 μl/min/nmol P450, respectively. NCLB was hydroxylated to 4′-hydroxydesmethylclobazam by CYP2C19 with Km = 5.74 μM, Vmax = 0.219 nmol/min/nmol P450, and CLint = 38.2 μl/min/nmol P450 (Hill coefficient = 1.54). These findings were supported by chemical inhibition studies in human liver microsomes. Indeed, ketoconazole (1 μM) inhibited the demethylation of CLB by 70% and omeprazole (10 μM) by 19%; omeprazole inhibited the hydroxylation of NCLB by 26%. Twenty-two epileptic patients treated with CLB were genotyped for CYP2C19. The NCLB/CLB plasma metabolic ratio was significantly higher in the subjects carrying one CYP2C19*2 mutated allele than in those carrying the wild-type genotype. CYP3A4 and CYP2C19 are the main P450s involved in clobazam metabolism. Interactions with other drugs metabolized by these P450s can occur; moreover, the CYP2C19 genetic polymorphism could be responsible for interindividual variations of plasma concentrations of N-desmethylclobazam and thus for occurrence of adverse events.
Clobazam (CLB) is a 1,5-benzodiazepine with antiepileptic properties, mainly used as adjuvant therapy in refractory epilepsy (Shorvon, 1995). Its major metabolic pathway is dealkylation to N-desmethylclobazam (NCLB), a pharmacologically active metabolite in which plasma concentrations are much higher than those of CLB in epileptic patients, suggesting that it may contribute to the therapeutic and adverse effects more than the parent compound does during long-term therapy (Jawad et al., 1984; Bardy et al., 1991; Shorvon, 1995). CLB is also metabolized into 4′-hydroxyclobazam (OH-CLB) and, finally, both NCLB and OH-CLB can be further converted to 4′-hydroxydesmethylclobazam (OH-NCLB) (Volz et al., 1979) (Fig. 1). The specific hepatic cytochrome P450 (P450) enzymes involved in CLB and NCLB metabolism have not been identified so far. Yet, some hypothesis can be made from the observation of metabolic interactions. Previous clinical studies have shown that conventional antiepileptic drugs can affect CLB metabolism. Particularly, inducers of P450 metabolism, mainly CYP2C/3A isoforms, such as phenytoin, carbamazepine, and phenobarbital, increase the clearance of CLB, leading to an accumulation of the N-desmethyl metabolite (Bardy et al., 1991; Sennoune et al., 1992; Theis et al., 1997). Concerning NCLB, a remarkably higher plasma concentration/dose ratio of NCLB and NCLB/CLB plasma concentration ratio could be observed in epileptic patients receiving comedication with felbamate, an in vitro inhibitor of CYP2C19 activity (Contin et al., 1999). Moreover, in subjects carrying defective CYP2C19*2 allele, Contin et al. (2002) observed higher NCLB/CLB plasma concentration/dose ratios than in subjects with no defective allele. These results suggest that CYP2C19 isoenzyme might be involved in NCLB metabolic clearance. Thus, because CLB is almost always used in association with other antiepileptic drugs, the risk of interaction is major and the knowledge of its metabolism would be very useful to explain and predict interactions. Therefore, the objective of this study was to identify and kinetically characterize in vitro the P450 isoforms responsible for the metabolism of CLB and its metabolites. In addition, 22 epileptic patients on stable CLB therapy were tested for CYP2C19*2 and *3 polymorphisms.
Major enzymatic pathways in the metabolism of clobazam. Clobazam undergoes either N-demethylation or C4′-hydroxylation to yield N-desmethylclobazam or 4′-hydroxyclobazam, respectively. Each of these metabolites can be further converted to 4′-hydroxydesmethylclobazam.
Materials and Methods
Chemicals and Reagents. Clobazam and N-desmethylclobazam were obtained from Laboratoires Roussel-Uclaf/Sanofi-Synthelabo France (Paris, France). 4′-Hydroxyclobazam and 4′-hydroxy-N-desmethylclobazam were synthesized and kindly provided by Laboratoires Biocodex (Montrouge, France).
Glucose 6-phosphate, glucose-6-phosphate dehydrogenase, nicotinamide adenine dinucleotide phosphate (NADP), 5-(p-hydroxyphenyl)-5-phenylhydantoin, ketoconazole, omeprazole, thiotepa, sulfaphenazole, quinidine, and chlorzoxazone were purchased from Sigma-Aldrich Chimie SARL. (St. Quentin Fallavier, France). Furafylline was obtained from Interchim (Montluçon, France).
Human Liver Microsomes and cDNA-Expressed P450 Isozymes. Human liver microsomes (cytomegalovirus-seronegative mixed gender pools) and specific human P450 enzymes (CYP1A1, -1A2, -2A6, -2B6, -2C8, -2C9, -2C18, -2C19, -2D6, -2E1, -3A4, -3A5, and -3A7) expressed in the Baculovirus-transfected insect cell system were purchased from BD Gentest (Woburn, MA). All cDNA-expressed P450 isozymes used also contain cDNA-expressed human P450 reductase and human cytochrome b5, these P450s having a higher activity than P450 without cytochrome b5. Microsomal protein concentrations and P450 contents were provided by BD Gentest.
Identification of the P450 Isoforms Involved in Clobazam Metabolism using cDNA-Expressed P450s. Incubation mixtures contained 100 mM phosphate buffer (pH 7.4), 0.5 mg · ml-1 MgCl2, 1 mM NADP+, 0.5 mg · ml-1 glucose 6-phosphate, 0.5 IU · ml-1 glucose-6-phosphate dehydrogenase, and the substrate (100 μM CLB, 14 μM NCLB, 72 μM OH-CLB) in a final incubation volume of 0.5 ml. The reactions were initiated by the addition of the P450 with a final P450 concentration of 50 nM, as recommended by BD Gentest. Incubations were performed for 30 min at 37°C and then stopped by the addition of 200 μl of ice-cold acetonitrile and cooling on ice. Incubations without NADPH-generating system served as controls. All incubations were conducted in duplicate.
Acetonitrile was chosen to dissolve CLB, NCLB, and OH-CLB, since it was shown to have the least inhibitory effect of a range of solvents (Chauret et al., 1998). It was present in incubation mixtures containing those compounds at a final concentration (v/v) of 0.4%. Due to their very limited solubility, 14 and 72 μM were the highest final concentrations that could be obtained for NCLB and OH-CLB, respectively.
Enzyme Kinetics with cDNA-Expressed CYP3A4, CYP2C19 and CYP2B6. Apparent Km and Vmax were determined for cDNA-expressed CYP3A4 and CYP2C19. CLB was incubated for 10 min with CYP3A4 and 30 min with CYP2C19 and CYP2B6 at 10 different concentrations (0.5, 1, 2, 5, 10, 20, 40, 60, 80, and 100 μM) for 3A4 and 2C19 and (10, 20, 40, 60, 80, 100, 200, 400, 600, 800, and 1000 μM) for 2B6. NCLB was incubated for 30 min with CYP2C19 at 10 different concentrations (0.5, 1, 1.5, 2, 4, 6, 8, 10, 12, and 14 μM).
Chemical Inhibition Studies with Human Liver Microsomes. Human liver microsomes and a P450-specific inhibitor (dissolved in acetonitrile) were added to an incubation mixture similar to that described above. The final concentration of total P450s was 200 nM. CLB and NCLB concentrations were chosen in the range of the therapeutic plasma concentrations (2 and 5 μM, respectively; Rey et al., 1999).
The P450-specific inhibitors used were 15 μM furafylline (CYP1A2 inhibitor), 10 μM thiotepa (CYP2B6 inhibitor), 1.5 μM sulfaphenazole (CYP2C9 inhibitor), 10 μM omeprazole (CYP2C19 inhibitor), 2 μM quinidine (CYP2D6 inhibitor), 5 μM chlorzoxazone (CYP2E1 inhibitor), and 1 μM ketoconazole (CYP3A4 inhibitor). They were used at concentrations corresponding to 5 times the Ki values found for each P450 isozyme-specific index reaction in human liver microsomes by Bourrie et al. (1996), except for omeprazole (Ko et al., 1997) and thiotepa (Rae et al., 2002).
High Pressure Liquid Chromatography (HPLC) Analysis.Determination of N-desmethylclobazam, 4′-hydroxyclobazam, and 4′-hydroxy-N-desmethylclobazam concentrations. The concentrations of the drugs were determined by a validated HPLC method. Briefly, the internal standard [5-(p-hydroxyphenyl)-5-phenylhydantoin] was added to the microsomal incubation mixture and the samples were extracted with 3 ml of tert-butylmethyl ether. After evaporation of the organic solvent, the residue was dissolved in the mobile phase (70:30 water/acetonitrile, 120 μl) and 85 μl were injected into the HPLC system (Thermo Finnigan, San Jose, CA). Separation was accomplished on an Ultrasphere ODS 5-μm 150 × 4.6 mm column (Beckman Coulter, Fullerton, CA) at a flow rate of 1 ml · min-1. An elution gradient was applied, beginning with 20% acetonitrile (Acn) and 80% water. After 20 min, Acn was increased to 30% and water decreased to 70% over 5 min. At 45 min, Acn was returned to 20% and water to 80%. The eluent was monitored by ultraviolet detection at a wavelength of 233 nm. Retention times for internal standard, OH-NCLB, OH-CLB, and NCLB were 12, 18, 27, and 33 min, respectively. The standard curves were linear from 2 to 1000 ng · ml-1 for NCLB and from 1 to 500 ng · ml-1 for OH-CLB and OH-NCLB. The coefficients of variation for interday reproducibility and precision were lower than 10% for the three metabolites.
Genotyping.Epileptic patients. The epileptic patients tested for CYP2C19 polymorphism participated in a randomized, placebo-controlled, add-on trial designed to test the efficacy of stiripentol in association with clobazam and valproate in severe myoclonic epilepsy in infancy. After a baseline period of 1 month, placebo or stiripentol was added to valproate and clobazam during a double-blind period of 2 months. Stiripentol or placebo was administered double-blind at the dose of 50 mg/kg/day. Maximum doses of clobazam and valproate were 0.5 and 30 mg/kg/day, respectively. Doses could be decreased by 25% for clobazam in case of drowsiness or hyperexcitability and by 10 mg/kg daily for valproate in case of loss of appetite. Minimum plasma concentrations of CLB and NCLB were measured at steady state of the baseline period using the HPLC method. CYP2C19 genotyping was performed on the archived plasma samples used for the CLB and NCLB plasma concentration assay. The study was approved by the local ethics committee (Pisa, Italy).
CYP2C19 genotyping. Total DNA was extracted from plasma samples using Qiamp DNA Blood Mini Kit (QIAGEN, Courtaboeuf, France). Genotyping for CYP2C19*2 and CYP2C19*3 was determined by polymerase chain reaction (PCR) followed by direct sequencing. PCR was performed according to a previously published method (De Morais et al., 1994) using a GenAmp PCR System 9700 (Applied Biosystems, Courtaboeuf, France). For CYP2C19*2 genotyping, the forward primer was AATTACAACCAGAGCTTGGC and the reverse primer TATCACTTTCCATAAAAGCAAG. For CYP2C19*3 genotyping, the forward primer was TATTATTATCTGTTAACTAATATGA and the reverse primer was ACTTCAGGGCTTGGTCAATA. Amplified DNA was purified using the QIAquick DNA purification system (QIAGEN) and sequenced using Big Dye-terminator chemistry and an ABI PRISM 3100 genetic analyzer (Applied Biosystems, Courtaboeuf, France).
Data Analysis. The percentages of metabolism for the expressed P450s were calculated using the ratio between the amount of metabolite formed and the amount of substrate initially introduced in the incubation mixture. The kinetics of CLB and NCLB biotransformations by cDNA-expressed CYP3A4 and CYP2C19 were fitted by a one-enzyme Michaelis-Menten model or a one-enzyme Hill model. Goodness of fit was based on visual examination of the plots and by application of the Akaike information criterion (Yamaoka et al., 1978). Calculated parameters were maximum rate of formation (Vmax), Michaelis constant (apparent Km), and intrinsic clearance (CLint = Vmax/apparent Km). Calculations were performed using Sigma Plot software (SPSS Inc., Chicago, IL). The percentages of inhibition were calculated by the ratio of the amounts of metabolites formed with and without the specific inhibitor.
Statistical Analysis. The comparison of metabolic ratio (NCLB/CLB), doses, and minimum plasma concentrations of CLB, NCLB, and valproate between patient subgroups was analyzed nonparametrically using the Mann-Whitney test (Prism 3.0; GraphPad Software Inc., San Diego, CA).
Results
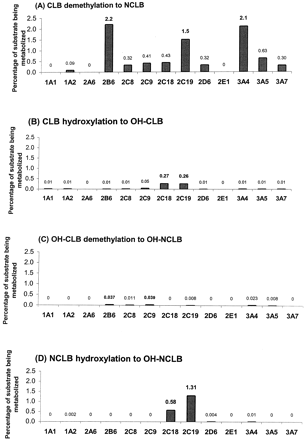
Identification of the P450 Isoforms Involved in Clobazam Metabolism using cDNA-Expressed P450s. Percentages of metabolism calculated after CLB incubation with the 13 different cDNA-expressed P450s suggested that CYP2B6 (2.2%), CYP3A4 (2.1%), and CYP2C19 (1.5%) mediated the formation of NCLB, whereas CYP2C18 (0.27%) and CYP2C19 (0.26%) mediated the formation of OH-CLB. NCLB was mainly metabolized by CYP2C19 (1.31%) and, at lower rates, by CYP2C18 (0.58%). OH-CLB was demethylated mainly by CYP2C9 (0.04%), CYP2B6 (0.04%), and CYP3A4 (0.02%) (Fig. 2).
Percentages of metabolism obtained for the 13 cDNA-expressed P450s tested in the four biotransformation pathways. Each substrate was incubated with each of the 13 cDNA-expressed P450s, and percentages of substrate being metabolized were calculated for the four biotransformation pathways. All incubations were done in duplicate. A, CLB to NCLB biotransformation; B, CLB to OH-CLB biotransformation; C, OH-CLB to OH-NCLB biotransformation; D, NCLB to OH-NCLB biotransformation.
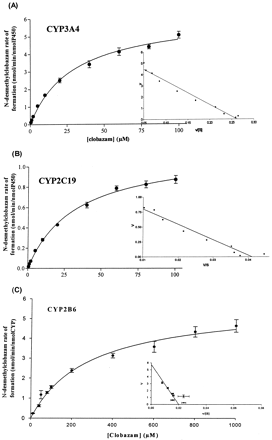
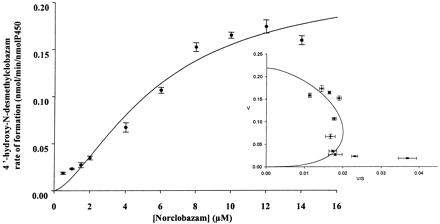
Kinetic Studies with cDNA-Expressed CYP3A4 and CYP2C19. The detailed kinetic analysis focused on the NCLB pathway, since the percentages of metabolism obtained for the demethylation pathway of CLB were higher than those obtained for the hydroxylation pathway (Table 1). The kinetic analysis focused on CYP3A4, CYP2C19, and CYP2B6. Under the experimental conditions used, the metabolism of CLB to NCLB by cDNA-expressed CYP3A4 was best described by a Michaelis-Menten model with a Km of 29.0 μM and a Vmax of 6.20 nmol/min/nmol P450 (Fig. 3A). The biotransformation of CLB to NCLB by cDNA-expressed CYP2C19 was best described by a Michaelis-Menten model with a Km of 31.9 μM and a Vmax of 1.15 nmol/min/nmol P450 (Fig. 3B). The biotransformation of CLB to NCLB by cDNA-expressed CYP2B6 was best described by a Michaelis-Menten model with a Km of 289 μM and a Vmax of 5.70 nmol/min/nmol P450 (Fig. 3C). The intrinsic clearance (CLint) of CLB N-demethylation was 6 and 11 times higher for CYP3A4 than that for CYP2C19 and CYP2B6, respectively (214 versus 36.1 and 19.7 μl/min/nmol P450). The hydroxylation of NCLB to OH-NCLB by cDNA-expressed CYP2C19 was also best described by a Hill model with a Km of 5.74 μM and a Vmax of 0.219 nmol/min/nmol P450 (Hill coefficient = 1.54) (Fig. 4).
Michaelis-Menten kinetic constants for the N-desmethylclobazam formation by cDNA-expressed CYP3A4, CYP2C19, and CYP2B6 and the 4′-hydroxy-N-desmethylclobazam formation by cDNA-expressed CYP2C19 Values are mean ± S.D.; each mean was obtained from three experiments in duplicate.
Mean (n = 6) rates of formation of N-desmethylclobazam from clobazam by cDNA-expressed CYP3A4 (A), CYP2C19 (B), and CYP2B6 (C). Inset, the data transformed by Eadie-Hofstee plot. Curves were fitted by a nonlinear regression one-enzyme Michaelis-Menten model. Each point represents the mean of three duplicates.
Mean (n = 6) rates of formation of 4′-hydroxy-N-desmethylclobazam from N-desmethylclobazam by cDNA-expressed CYP2C19. Inset, the data transformed by Eadie-Hofstee plot. Curves were fitted by a nonlinear regression one-enzyme Hill model. Each point represents the mean of three duplicates.
Chemical Inhibition Studies. Inhibition of CLB and NCLB metabolism in human liver microsomes was performed using the same CLB and NCLB concentrations as those used for the kinetic studies (2 and 5 μM, respectively). CLB and NCLB biotransformations were linear with respect to incubation time and to P450 concentration in microsomes.
Ketoconazole (1 μM) decreased NCLB formation by 69.7 ± 6.6%, and omeprazole (10 μM) and thiotepa (10 μM) reduced it by 18.7 ± 8.8% and 24.3 ± 8.0%, respectively, whereas sulfaphenazole (1.5 μM), furafylline (15 μM), chlorzoxazone (5 μM), and quinidine (2 μM) decreased it by less than 8%. These results confirm the roles played by CYP3A4, CYP2C19, and CYP2B6 in the demethylation of CLB. NCLB hydroxylation was mainly inhibited by omeprazole (25.9 ± 1.5%), confirming the major role of CYP2C19, and less by ketoconazole (12.3 ± 2.8%). Results are presented in Table 2.
Percentages of inhibition for CLB (2 μM) demethylation and NCLB (5 μM) hydroxylation measured in human liver microsomes in the presence of specific inhibitors. Values are mean ± S.D.
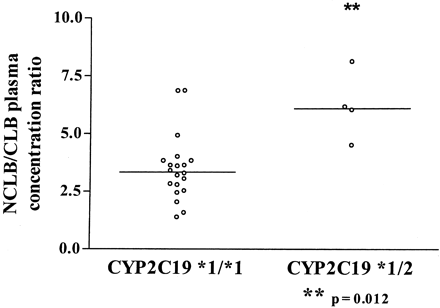
CYP2C19 Genotyping.Epileptic patients. Twenty-two epileptic children aged 3.5 to 18.9 years (means 8.7 years) were included in the study. Two CYP2C19 genotypes were identified in the patients studied: *1/*1 (n = 18) and *1/*2 (n = 4). The CYP2C19*3 variant was not observed. The NCLB/CLB metabolic ratio was significantly higher in the subjects carrying the *2 allele (CYP2C19*1/*2) than in the *1/*1 subjects (p = 0.012) (Fig. 5). Mean clobazam and valproate doses and mean minimum plasma concentration are presented in Table 3.
In vivo N-desmethylclobazam to clobazam plasma concentration ratio segregated according to CYP2C9 genotype. Genotyping was done in epileptic children (n = 22, aged from 3.5 to 18.9 years) all receiving clobazam and valproate as comedications. NCLB/CLB plasma concentration ratio was significantly higher (p = 0.012) in CYP2C19*1/*2 than in CYP2C19*1/*1 subjects. The bar represents the median value of the distribution.
Doses of CLB and valproate, minimum plasma concentrations of CLB, NCLB, and valproate, and NCLB/CLB plasma metabolic ratios (mean ± S.D. (median) or [range]) in epileptic children 3.5 to 18.9 years old, treated with clobazam and valproate as comedications
Discussion
For the first time, the main metabolic pathways of clobazam were characterized. Our data using Baculovirus-expressed P450s strongly suggest that CLB demethylation to NCLB in the human liver is primarily mediated by CYP3A4 and CYP2C19, CLB hydroxylation seems to be a minor metabolism pathway mainly mediated by CYP2C18 and CYP2C19. Indeed, the percentages of metabolism obtained with the expressed P450s were much higher for the demethylation pathway (≥1.5%) than for the hydroxylation pathway (<0.3%). This is consistent with in vivo observations indicating that the demethylation pathway may be of greater importance than the hydroxylation route for CLB metabolism, since patients treated with CLB have high NCLB plasma concentration, whereas they have undetectable OH-CLB plasma concentrations (unpublished data from our laboratory). In human liver, CYP3A4 and CYP2C19 account, respectively, for about 30% (Shimada et al., 1994) and 1.4% (Inoue et al., 1997) of total hepatic microsomal P450, thus, suggesting that CYP3A4 is the major contributor to the CLB demethylation pathway. These results are also supported by the chemical inhibition studies, since ketoconazole inhibited by 70% the NCLB formation in human liver microsomes. Concerning the hydroxylation of NCLB to OH-NCLB, expressed P450s and chemical inhibition studies both demonstrated the major involvement of CYP2C19, with a percentage of substrate being metabolized comparable to that obtained for NCLB formation (>1%), suggesting the importance of this pathway in the in vivo OH-NCLB formation compared with the demethylation of OH-CLB that seems to be a minor pathway mediated by CYP2B6, CYP2C9, and CYP3A4, with percentages of metabolism lower than 0.05%. In addition, despite the fact that the percentage of substrate being metabolized obtained with CYP2C18 for the NCLB hydroxylation is not negligible (0.58%), this pathway probably must not be clinically relevant since the CYP2C18 protein is not expressed at a significant level in human liver (Richardson et al., 1997; Gerbal-Chaloin et al., 2001). Thus, the identification of CYP2C19 as the major P450 involved in the NCLB biotransformation is consistent with the accumulation of this metabolite in epileptic patients receiving felbamate cotherapy (Contin et al., 1999), since felbamate inhibits CYP2C19 and, then, the elimination of NCLB. These results are also consistent with studies concerning the metabolism of several 1,4-benzodiazepines that have shown the major role mediated by the CYP3A4 and CYP2C19 isoforms: flunitrazepam (Hesse et al., 2001; Kilicarslan et al., 2001), diazepam (Jung et al., 1997; Yang et al., 1998), and midazolam and triazolam (Perloff et al., 2000). Indeed, considering the structural analogy between the 1,4- and 1,5-benzodiazepines, the same P450 isoforms were likely to be involved.
Substrate activation was observed for hydroxylation of NCLB by CYP2C19. Sigmoidal enzyme kinetics had previously been reported by Venkatakrishnan et al. (1998) for adinazolam N-demethylation by human recombinant CYP2C19. This result suggests cooperative kinetics. However, further experiments are needed to confirm this hypothesis.
The percentage of inhibition obtained with thiotepa (24%) is similar to that obtained with omeprazole. This finding could be attributable to the similar levels of activity of CYP2B6 and CYP2C19 in the microsomes used (47 and 40 pmol/mg/min, respectively), However, we conclude that the involvement of CYP2B6 in CLB demethylation must be much lower than that of CYP3A4 and CYP2C19 regarding its very low amount in the human liver [0.2% versus about 30% (Shimada et al., 1994) and 1.4% (Inoue et al., 1997) in Caucasians, respectively]. In addition, Hamaoka et al. (2001) showed the minor involvement of this P450 in midazolam metabolism in human liver microsomes despite a significant activity of biotransformation obtained with expressed P450s. Yet, it is interesting to note that CYP2B6 is able to demethylate CLB to NCLB since it is inducible by drugs such as phenobarbital or rifampin (Faucette et al., 2004).
Because CLB and its active metabolite NCLB are metabolized by CYP2C19, the polymorphic expression of this enzyme may affect the clinical safety and efficacy of this drug. Indeed, CYP2C19 is genetically polymorphic, with large differences in the frequency of the poor metabolizer phenotype among different populations [18-23% of Chinese and 2-5% of Caucasians are homozygous for deficient CYP2C19 alleles and lack enzyme activity (De Morais et al., 1994)], and this polymorphism affects the metabolism of some commonly prescribed drugs such as omeprazole, propranolol, and the psychotropic drugs diazepam, citalopram, sertraline, and imipramine (Poolsup et al., 2000; Goldstein, 2001). Concerning CLB, one study has provided evidence that subjects carrying one or two copies of the defective CYP2C19*2 allele might develop markedly elevated steady-state plasma concentrations of NCLB and be at higher risk of adverse effects, principally sedation, dizziness, and fatigue (Contin et al., 2002). The potential clinical importance of the CYP2C19 genetic polymorphism has also already been tested for another benzodiazepine, flunitrazepam, in a small pilot study in which individuals who lacked CYP2C19 activity exhibited higher plasma flunitrazepam concentrations and demonstrated greater sedation and psychomotor impairment (Kilicarslan et al., 2001). Like Contin et al. (2002), we observed that subjects carrying one CYP2C19*2 allele had NCLB/CLB plasma concentration ratios higher than subjects with no mutant allele, suggesting a gene-dosage effect with CYP2C19. This result was in accordance with previous published works on other CYP2C19 substrates such as diazepam (Qin et al.,1999), omeprazole (Lamba et al.,2001), fluoxetine (Liu et al., 2002), or carisoprodol (Bramness et al., 2003).
In the in vivo study, all patients were receiving valproate in addition to clobazam. The difference observed for the metabolic ratio (NCLB/CLB) between CYP2C19*1/*1 and CYP2C19*1/*2 subjects could not be due to an influence of valproate on CYP2C19, since the main metabolic pathways for valproate are conjugation with glucuronic acid and mitochondrial β-oxidation (DeVane, 2003). Furthermore, valproate has not been shown to inhibit CYP2C19 significantly (Wen et al., 2000).
The recognized contribution of CYP2C19 to NCLB metabolism might also help to explain a previous report of phenytoin toxicity in patients receiving add-on CLB, possibly due to interference with the CYP2C19 component of phenytoin metabolism (Zifkin et al., 1991). Moreover, the potential interaction of CLB with other antiepileptic drugs, such as diazepam, a substrate of CYP2C19 (Jung et al.,1997), or topiramate, an inhibitor of CYP2C19 (Levy et al., 1995), might be expected.
In summary, this study showed that CYP3A4 and CYP2C19 are the main enzymes involved in the N-demethylation of CLB to NCLB and that CYP2C19 is the major contributor to the hydroxylation of NCLB to OH-NCLB. The polymorphic expression of CYP2C19 might influence the clinical safety and efficacy of CLB.
Acknowledgments
We are indebted to M. Cammas for excellent technical assistance.
Footnotes
-
This work was supported by a grant from Laboratoires Biocodex (Montrouge, France).
-
ABBREVIATIONS: CLB, clobazam; NCLB, N-desmethylclobazam; OH-CLB, 4′-hydroxyclobazam; OH-NCLB, 4′-hydroxydesmethylclobazam; P450, cytochrome P450; HPLC, high performance liquid chromatography; Acn, acetonitrile; PCR, polymerase chain reaction; CLint, intrinsic clearance.
- Received January 5, 2004.
- Accepted July 30, 2004.
- The American Society for Pharmacology and Experimental Therapeutics