Abstract
Induction of cytochrome P450 3A4 (CYP3A4) is determined typically by employing primary culture of human hepatocytes and measuring CYP3A4 mRNA, protein and microsomal activity. Recently a pregnane X receptor (PXR) reporter gene assay was established to screen CYP3A4 inducers. To evaluate results from the PXR reporter gene assay with those from the aforementioned conventional assays, 14 drugs were evaluated for their ability to induce CYP3A4 and activate PXR. Sandwiched primary cultures of human hepatocytes from six donors were used and CYP3A4 activity was assessed by measuring microsomal testosterone 6β-hydroxylase activity. Hepatic CYP3A4 mRNA and protein levels were also analyzed using branched DNA technology/Northern blotting and Western blotting, respectively. In general, PXR activation correlated with the induction potential observed in human hepatocyte cultures. Clotrimazole, phenobarbital, rifampin, and sulfinpyrazone highly activated PXR and increased CYP3A4 activity; carbamazepine, dexamethasone, dexamethasone-t-butylacetate, phenytoin, sulfadimidine, and taxol weakly activated PXR and induced CYP3A4 activity, and methotrexate and probenecid showed no marked activation in either system. Ritonavir and troleandomycin showed marked PXR activation but no increase (in the case of troleandomycin) or a significant decrease (in the case of ritonavir) in microsomal CYP3A4 activity. It is concluded that the PXR reporter gene assay is a reliable and complementary method to assess the CYP3A4 induction potential of drugs and other xenobiotics.
Human CYP3A4 is the single most important drug-metabolizing enzyme. It accounts for roughly 40% of the total cytochrome P450 in human liver microsomes and metabolizes more than 50% of the clinically used drugs (Lehmann et al., 1998). CYP3A4 is also expressed in the small intestine, where it contributes to the first-pass elimination of many drugs (Paine et al., 1996). The hepatic and intestinal expression of CYP3A4 can be induced in vivo by a variety of drugs, such as rifampin and phenobarbital, leading to the accelerated metabolism of the drugs themselves and concomitantly used drugs. Consequently, the therapeutic effects of drugs may be altered as a result of CYP3A4 induction (Pichard et al., 1990). Because of the potential for induction-based drug-drug interactions, it is important to screen all new chemical entities for their CYP3A4 induction potential.
Generally, CYP3A4 induction is assessed in vitro by treating primary cultures of human hepatocytes with test compound, preparing microsomes from the treated hepatocytes, and measuring the 6β-hydroxylation of testosterone (a selective marker of CYP3A4 activity) (Li et al., 1992;Pearce et al., 1996; LeCluyse et al., 2000). Alternatively, CYP3A4 induction can be assessed at the protein level or at the mRNA level. This approach has certain disadvantages, such as, the considerable interindividual donor variation in response to the CYP3A4 inducers. Cell culture conditions are also an important factor and may be the cause of considerable variability (LeCluyse et al., 2000, 2001). For instance, the active cytochrome P450 and phase II enzymes in cultured hepatocytes could metabolize the test compound and affect the induction results. Many of the aforementioned variables can be controlled or accounted for on a case-by-case basis. However, a consistent disadvantage of the human hepatocyte culture model continues to be the availability and quality of donor tissue, which dictates the number and timing of in vitro induction studies.
Even though CYP3A4 induction has been known for decades, the molecular mechanism of induction remained an enigma until recently. PXR1, also called steroid and xenobiotic receptor or pregnane-activated receptor, was found to mediate the drug-induced expression of CYP3A4 (Bertilsson et al., 1998; Lehmann et al., 1998; Xie et al., 2000). Based on this mechanism of induction, a cell-based PXR reporter gene assay has been established to screen CYP3A4 inducers (Lehmann et al., 1998; Goodwin et al., 1999; Moore et al., 2000). The PXR reporter gene assay has some advantages over primary cultures of human hepatocytes. For example, the reporter gene assay uses inexpensive human-derived cell lines, such as the hepatocellular carcinoma HepG2. Cell lines are also more amenable to establishing automated high-throughput formats for screening the induction potential of new drugs. However, further comparison between this new procedure and the conventional method based on primary cultures of human hepatocytes is still necessary. Therefore, in the present study, 14 commercially available drugs were studied with respect to their capacity to activate PXR in the reporter gene assay and induce CYP3A4 mRNA, protein, and activity in primary cultures of human hepatocytes.
Experimental Procedures
Materials.
Dexamethasone-t-butylacetate was purchased from Research Plus, Inc. (Bayonne and Denville, NJ). Phenobarbital was purchased from Amend Drug and Chemical Company (Irvington, NJ). Ritonavir was obtained from Abbott Laboratories (Abbott Park, IL). Sulfinpyrazone was purchased from ICN Pharmaceuticals Biochemicals Division (Aurora, OH). Carbamazepine, clotrimazole, dexamethasone, methotrexate, phenytoin, probenecid, rifampin (rifampicin), sulfadimidine (sulfamethazine), taxol (paclitaxel), and troleandomycin were purchased from Sigma-Aldrich (St. Louis, MO). Testosterone was purchased from Steraloids, Inc. (Wilton, NH).
PXR Cloning and Expression Construction.
A full-length PXR open reading frame was amplified by RT-PCR from human liver RNA using gene-specific primers 5′- AACCTGGAGGTGAGACCCAAAGA-3′ and 5′-ATCTCGAGGATCCTCAGCTACCTGTGATGCCGA-3′ (Bertilsson et al., 1998;Lehmann et al., 1998). The resulting amplicon was digested withEcoRI and XhoI and subcloned into the polylinker site of pCDNA3 (Invitrogen, Carlsbad, CA) for expression, hereafter designated pCDNA3-hPXR. Correct PXR sequence and insert orientation were confirmed by DNA sequence analysis.
Construction of pGL3–3A4 Reporter.
Chirmeric CYP3A4 luciferase reporter vector was prepared following the method described previously (Goodwin et al., 1999). The resulting construct contained two fragments of CYP3A4 5′-flanking region (−7836 to −7208 and −362 to +53) linked to luciferase reporter sequence of pGL3-basic vector (Promega, Madison, WI) and hereafter designated pGL3–3A4. Correct insert orientation was confirmed by DNA sequence analysis.
PXR Reporter Gene Assay.
HepG2/C3A cells (American Type Culture Collection, Manassas, VA) were plated in 96-well plates (Packard Bioscience, Meriden, CT) (2 × 104 cells per well) in 100 μl of plating medium (phenol-red-free Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (charcoal stripped), 2 mMl-glutamine and 1 mM nonessential amino acids) and incubated overnight. Following the incubation, the cells were transfected by lipofection using Fugene-6 (Roche Applied Science (Indianapolis, IN) in the presence of serum following the manufacturer's instruction. Transfection mixes applied to each well contained 2.5 ng of hPXR expression vector pCDNA3-hPXR, 50 ng of luciferase reporter plasmid pGL3–3A4, and 20 ng of alkaline phosphatase expression vector pSEAP2-Control (BD Biosciences Clontech, Palo Alto, CA). The media was replaced with 100 μl of designed fresh dosing medium (plating medium + test articles in 0.25% DMSO) 6 h following transfection and incubated for an additional 42 h. All test articles except phenobarbital were tested at final concentrations of 2, 10, 20, and 50 μM. For reasons outlined later, phenobarbital was tested at 20, 50, 150, and 250 μM. The reporter activities of firefly luciferase and alkaline phosphatase were determined by Steady-Glo luciferase assay system (Promega) and GreatEscAPe chemiluminescence detection kit (BD Biosciences Clontech), respectively, according to the suppliers' specifications.
Primary Culture of Human Hepatocytes.
Human liver tissues were obtained as surgical wastes or rejected donor livers from the University of North Carolina at Chapel Hill School of Medicine or from the Department of Surgery at the University of Kansas Medical Center and National Disease Research Interchange (Philadelphia, PA). All tissues were obtained through qualified medical staff, with donor consent and with the approval of the appropriate hospital ethics committees. Clinical characteristics of liver donors are summarized in Table 1. Human hepatocytes were isolated by a modification of the two-step collagenase digestion method (Li et al., 1992), with a minor modification (LeCluyse et al., 2000). The isolated human hepatocytes were cultured for CYP3A4 induction studies as described previously (Li et al., 1992; LeCluyse et al., 2000). Human hepatocytes from liver donors HL-N095, HL-N098, HL-N100, H249, and H254 were used to test all 14 compounds. Hepatocytes from donor H232 were used only for dexamethasone, dexamethasone-t-butylacetate, methotrexate, and phenobarbital; hepatocytes from donor H233 were used for the 10 remaining test compounds. Primary cultures of human hepatocytes were maintained for 2 days followed by 3 daily doses of test compounds at a final concentration of 2, 10, and 20 μM (except for phenobarbital, which was tested at 50, 150, and 250 μM). The final concentration of DMSO in culture medium was 0.1% (v/v), and the vehicle control group received only 0.1% DMSO.
Clinical information on liver donors
Testosterone Oxidation Assay.
Microsomes were prepared from the cultured human hepatocytes 24 h after the final drug treatment as described previously (Li et al., 1992; LeCluyse et al., 2000). Concentration of protein was determined using bicinchoninic acid protein assay (Pierce Chemical Co., Rockford, IL). Microsomal CYP3A4 activity was determined by measuring the 6β-hydroxylation of testosterone (Pearce et al., 1996). CYP3A4 induction was expressed as -fold induction over the vehicle control.
Branched DNA Signal Amplification Technology.
Specific oligonucleotide probe (blockers, capture extenders, and label extenders) sets for bDNA analysis of CYP3A4 mRNA were developed by XenoTech LLC (Kansas City, KS) (Czerwinski et al., 2002) and synthesized commercially. CYP3A4 mRNA levels in cultured human hepatocytes were examined as described by Czerwinski et al. (2002) and are expressed as a ratio of glyceraldehyde 3-phosphate dehydrogenase mRNA levels to correct for well-well variation.
Northern Blot.
Total RNA was prepared from cultured hepatocytes of liver donor HL-N100 using Trizol reagent (Invitrogen), and 10 μg of total RNA was blotted to GeneScreenPlus membrane (PerkinElmer Life Sciences, Boston, MA). A 389 base pair EcoRI fragment of human CYP3A7 from plasmid pHFLA-A (kindly provided by Dr. Erin Schuetz at St. Jude Children's Research Hospital, Memphis, TN) was used to probe CYP3A4 mRNA (Schuetz et al., 1993). This probe is able to identify all the human CYP3A members because of high identity among the human CYP3A family members (Domanski et al., 2001). Since CYP3A5 and 3A43 (a newly discovered CYP3A member) are expressed in hepatocytes at extremely low level (Schuetz et al., 1993; Domanski et al., 2001), and the fetal specific gene CYP3A7 is expressed at low levels in adult hepatocytes in primary culture (Greuet et al., 1996), the results obtained from Northern blot analysis with this probe should predominantly reflect CYP3A4 mRNA. In a minority of white and Chinese people, CYP3A5 is polymorphically expressed at high levels, representing at least 50% of the total hepatic CYP3A content in people with at least one CYP3A5*1 allele (Chou et al., 2001; Kuehl et al., 2001). The mRNA was quantitated by scanning densitometry of the autoradiographs and normalized against β-actin mRNA.
Immunoblot Analysis.
Equal amounts of microsomal protein (10 μg) were loaded onto polyacrylamide gels and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Levels of immunoreactive CYP3A4 protein in human hepatocyte microsomes were determined by Western immunoblotting analysis with CYP3A4-specific antibodies at a 1:2000 dilution (Chemicon International Inc., Temecula, CA) as described previously (LeCluyse et al., 2000). The relative amounts of CYP3A4 protein were estimated from densitometric analysis of the blot after scanning.
Statistics.
Differences between two groups were analyzed using two-sided two-samplet test. Correlations were measured by Pearson's correlation coefficient (r) and Spearman's Rho (ρ) (Conover, 1980).
Results
PXR Activation Using a Reporter Gene Assay.
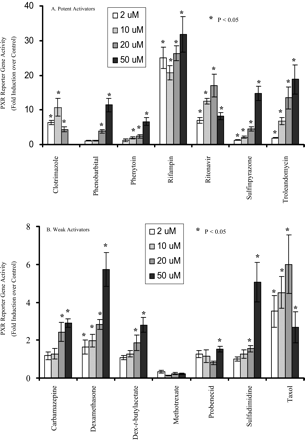
As shown in Fig. 1, PXR was activated strongly (>10-fold) by rifampin, ritonavir, troleandomycin, sulfinpyrazone, phenobarbital (250 μM), and clotrimazole; moderately (5- to 10-fold) by phenytoin, taxol, dexamethasone and sulfadimidine; and weakly (<5-fold) by carbamazepine and dexamethasone-t-butylacetate. Methotrexate and probenecid did not show dose-dependent PXR activation at the concentrations tested. Results obtained from previous PXR reporter gene assays (Bertilsson et al., 1998; Lehmann et al., 1998; Goodwin et al., 1999;Harvey et al., 2000; Moore et al., 2000) performed on dexamethasone, dexamethasone-t-butylacetate, clotrimazole, phenobarbital, and rifampin are in general agreement with the present findings.
PXR activation by test compounds in the reporter gene assay.
A, potent activators; B, weaker activators. HepG2/C3A cells were seeded on 96-well plates on day 1, transfected with pCDNA3-hPXR, pGL3–3A4, and pSEAP2-Control for 6 h on day 2, and treated with test compounds for 42 h. The reporter activity of firefly luciferase was determined on day 4. The results are the mean of six determinations. Phenobarbital's concentrations were 20, 50, 150, and 250 μM, respectively. In this and all the other figures, dexamethasone-t-butylacetate is abbreviated as dex-t-butylacetate. Differences were analyzed by two-sided, two-sample t test. ★, P< 0.05 compared with vehicle control.
CYP3A4 Activity in Cultured Human Hepatocytes.
As shown in Table 2, the basal activity of CYP3A in microsomes from the seven cultured human hepatocytes differed more than 8-fold. The induction of CYP3A activity by rifampin at 20 μM ranged from 2-fold (HL-N098) to nearly 10-fold (H254). It appears that the human hepatocytes with lower basal activity exhibit a greater -fold induction of CYP3A4 activity, possibly due to the limited maximal capacity of CYP3A4 expression in human hepatocytes and/or negative feedback regulation by CYP3A4 itself (Schuetz and Strom, 2001).
Interindividual variation in CYP3A4 basal activity and induction by rifampin in primary cultures of human hepatocytes
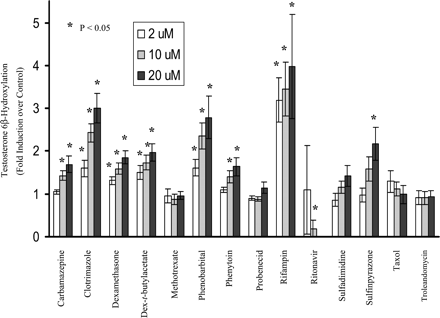
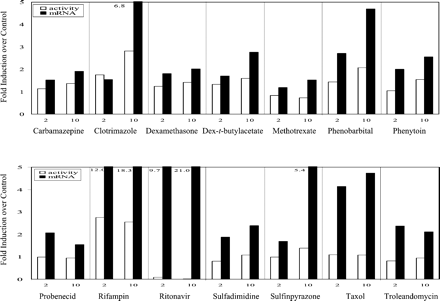
Induction of CYP3A4 in primary cultures of human hepatocytes by the 14 compounds is shown in Fig. 2. CYP3A4 activity was induced strongly by rifampin and clotrimazole (3-fold or higher); moderately by phenobarbital (250 μM) and sulfinpyrazone (2- to 3-fold); and weakly by carbamazepine, dexamethasone, dexamethasone-t-butylacetate, and phenytoin (1.5- to 2-fold). Methotrexate, probenecid, sulfadimidine, taxol, and troleandomycin failed to induce CYP3A4 activity. Notably, CYP3A4 activity was significantly decreased by ritonavir at final concentrations of 10 and 20 μM. However, at a final concentration of 2 μM, ritonavir did induce hepatic CYP3A4 activity 6-fold over the control in one of six liver donors (H254).
Induction of microsomal CYP3A4 activity by test compounds in primary culture of human hepatocytes.
Hepatocyte cultures were treated for 3 days with the test compounds at 2, 10, and 20 μM, respectively, except for phenobarbital, which was dosed at 50, 150, and 250 μM, respectively. DMSO at 0.1% (v/v) was used as vehicle control. Microsomes were prepared 24 h after the last treatment, and the CYP3A4 activity was determined by measuring testosterone 6β-hydroxylase activity (pmol/mg/min). The results are the mean ± standard errors of six experiments from seven donors as described under Experimental Procedures. Differences were analyzed by two-sided, two-sample t test ★,P < 0.05 compared with control.
The induction results obtained from the present study are consistent with those reported previously with carbamazepine, dexamethasone, phenobarbital, phenytoin, rifampin, and sulfinpyrazone (Pichard et al., 1990; LeCluyse et al., 2000). In the present study, sulfadimidine and troleandomycin failed to markedly induce CYP3A4 activity in human hepatocytes, and dexamethasone induced CYP3A4 activity only weakly, probably due to the low concentrations used in these studies. Taxol at a concentration of 2 μM showed 1.8- and 2.3-fold induction of CYP3A4 activity in donors H249 and H254, respectively, but less than 1-fold in all other donors. Methotrexate and probenecid, two metabolically stable xenobiotics in vivo that have not been reported to induce CYP3A4, were chosen as negative controls in the present study. As expected, they did not alter the CYP3A4 activity at the concentrations tested, although the plasma concentrations of probenecid in patients may be much higher than 20 μM. Ritonavir is a known CYP3A4 substrate and inhibitor (Hsu et al., 1998a). Clinical studies have indicated that it induces CYP3A4 after administration to healthy volunteers (Gass et al., 1998; Ouellet et al., 1998) or to patients (Geletko and Erickson, 2000), although there are no reports that ritonavir induces CYP3A4 in primary cultures of human hepatocytes.
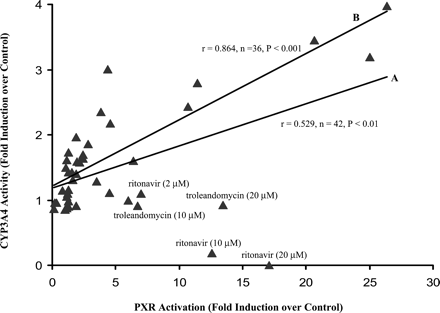
The correlation between PXR activation from the reporter gene assay and CYP3A4 activity from primary cultures is relatively poor, with a correlation coefficient r = 0.529 (or ρ = 0.426,n = 42, P < 0.01), as shown in Fig.3. In the absence of two outliers, ritonavir and troleandomycin, the correlation between the two assays was markedly improved, r = 0.864 (or ρ = 0.831,n = 36, P < 0.001), as also shown in Fig. 3.
Correlation between the PXR reporter gene assay and testosterone 6β-hydroxylation assay.
PXR activation by test compounds was obtained in the reporter gene assay, as shown in Fig. 1. Induction of CYP3A4 activity by test compounds was obtained in human hepatocytes of six donors, as shown in Fig. 2. For the correlation analysis, only the paired results generated in the two assays at the concentrations of 2, 10, and 20 μM were employed. Correlations were measured by Pearson's correlation coefficient (r) and Spearman's Rho (ρ). (A) in the presence of all 14 test compounds, and (B) in the absence of ritonavir and troleandomycin.
CYP3A4 mRNA Levels in Cultured Human Hepatocytes.
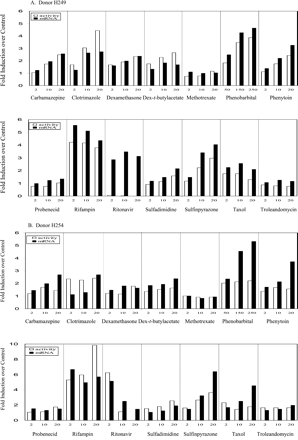
CYP3A4 mRNA levels in cultured hepatocytes were determined using a bDNA assay for liver donors H249 and H254 with treatment of test compounds at all concentrations (Fig. 4) and using Northern blot analysis for donor HL-N100 at 2 and 10 μM of all test compounds (Fig. 5). Overall, CYP3A4 mRNA expression was induced strongly by rifampin, sulfinpyrazone, ritonavir, and clotrimazole; moderately by phenytoin, taxol, and phenobarbital (250 μM); and weakly by carbamazepine, dexamethasone, and dexamethasone-t-butylacetate but not by methotrexate, probenecid, sulfadimidine, and troleandomycin.
CYP3A4 mRNA levels in primary cultures of human hepatocytes determined by bDNA assay.
A, liver donor H249; B, liver donor H254. Cultured hepatocytes were treated as described in Fig. 2 prior to adding RNA isolation buffer. CYP3A4 mRNA levels were determined by bDNA amplification assay and normalized against glyceraldehyde-3-phosphate dehydrogenase mRNA. The results are the mean of three determinations. For a comparison, the respective CYP3A4 activities (6β-hydroxylation of testosterone) from the same donor are shown as the open bars.
CYP3A4 mRNA levels in primary culture of human hepatocytes determined by Northern blotting.
Total RNA was prepared from cultured hepatocytes of liver donor HL-N100 using Trizol reagent, and 10 μg of total RNA was blotted to GeneScreenPlus membrane. A 389 base pair EcoRI fragment of human CYP3A7 from plasmid pHFLA-A was used as probe for CYP3A4. The CYP3A4 mRNA levels were estimated from densitometric analysis of the blot after scanning and normalized against β-actin mRNA. For comparison, the respective CYP3A4 activities (6β-hydroxylation of testosterone) from the same donor are shown as the open bars.
A modest correlation was observed between PXR activation and induction of CYP3A4 mRNA levels. In the donors H249 and H254 (mRNA levels obtained using bDNA assay), correlation coefficient is 0.634 (or ρ = 0.650, n = 84, P < 0.001), as shown in Fig. 6. Also, CYP3A4 mRNA levels generally correlated well with the respective microsomal CYP3A4 activities, with the exception of ritonavir. In the donors H249 and H254, correlation coefficient is 0.706 (n = 84,P < 0.001, correlation figure not shown). Ritonavir activated the transcription of CYP3A4 in the cultured human hepatocytes at all concentrations tested, but decreased the respective microsomal CYP3A4 activity, compared with the vehicle controls.
Correlation between the PXR activation and induction of CYP3A4 mRNA level.
PXR activation by test compounds was obtained in the reporter gene assay, as shown in Fig. 1. Induction of CYP3A4 mRNA level by test compounds was obtained in human hepatocytes of donors H249 and H254, as shown in Fig. 4. For the correlation analysis, only the paired results generated in the two assays at the concentrations of 2, 10, and 20 μM were employed. Correlations were measured by Pearson's correlation coefficient (r) and Spearman's Rho (ρ).
CYP3A4 Protein Levels in Cultured Human Hepatocytes.
Microsomal CYP3A4 protein levels from hepatocyte cultures prepared from HL-N095 were measured by Western immunoblotting (Fig.7A). For a better comparison, the blot was scanned, digitized, and the -fold increases over the vehicle controls plotted (Fig. 7B). Clotrimazole, phenobarbital, phenytoin, rifampin, and sulfinpyrazone all increased the levels of CYP3A4 immunoreactive protein, whereas methotrexate and probenecid did not show any induction. The CYP3A4 protein levels highly correlated with the CYP3A4 activities present in the same donor (r = 0.832, n = 42, P < 0.001). In the absence of ritonavir, the correlation is slightly increased (r = 0.892, n = 39, P< 0.001). Ritonavir moderately increased CYP3A4 protein levels, but the respective CYP3A4 protein bands were somewhat smeared, suggesting the covalent binding of drug to the protein. It appeared that troleandomycin did not increase CYP3A4 protein levels markedly, being less than 130% of the control at the concentrations tested.
Induction of immunoreactive CYP3A4 levels in cultured human hepatocytes.
A, equal amounts of microsomal protein (10 μg) from donor HL-N095 were used for Western blot analysis with CYP3A4-specific antibodies. B, the blots were scanned and the digital data were generated. For comparison, the respective CYP3A4 activity (6β-hydroxylation of testosterone) from the same donor is shown (the open bar).
Discussion
Induction of CYP3A4 has been shown to alter the pharmacokinetics of many drugs, for which reason in vitro procedures have been developed to screen new chemical entities and drug candidates as CYP3A4 inducers. Typically, the potential of test compounds to induce CYP3A4 is examined in vitro in primary cultures of human hepatocytes by measuring changes in the metabolism of probe substrates for CYP3A4, such as testosterone and midazolam, or by measuring changes in the levels of CYP3A4 mRNA and protein. Recently, a reporter gene assay was established to identify compounds that activate PXR, the receptor implicated in drug-induced CYP3A4 gene expression. While the reporter gene assay has certain advantages over the conventional hepatocyte culture system, additional studies are required to demonstrate the validity of this newer approach. The present study was undertaken to compare the PXR reporter gene assay and the primary cultures of human hepatocyte system for measuring CYP3A4 induction.
Fourteen commercially available compounds were evaluated for their ability to induce CYP3A4 in hepatocytes and activate PXR in a reporter gene assay. Most of the test compounds have been reported to cause low, moderate, or high induction of CYP3A4 in vitro and in some cases in vivo (Pichard et al., 1990; Gass et al., 1998; Kostrubsky et al., 1998,1999; Lehmann et al., 1998; Ouellet et al., 1998; Goodwin et al., 1999;Geletko and Erickson, 2000; LeCluyse et al., 2000; Ledirac et al., 2000). Ritonavir was reported to induce CYP3A4 in healthy volunteers or patients after 10 to 16 days administration (Hsu et al., 1997; Gass et al., 1998; Ouellet et al., 1998; Geletko and Erickson, 2000). Thus far, there are no reports of the induction of CYP3A4 by ritonavir in cultured human hepatocytes. In humans, ritonavir causes reversible and irreversible CYP3A4 inhibition, and most of the drug-drug interactions reported for ritonavir reflect its inhibitory effect. For example, treatment with ritonavir markedly increases the area under the plasma concentration-time curve and peak concentration of saquinavir (50- and 22-fold, respectively) (Hsu et al., 1998b). Methotrexate and probenecid were chosen as negative controls because they are not significantly metabolized in vivo and because there are no reports implicating them as CYP3A4 inducers.
High interindividual variability in CYP3A4 expression and inducibility has been observed in primary cultures of human hepatocytes. In the present study, basal CYP3A activity varied approximately 8-fold, and the magnitude of induction by the positive control rifampin ranged from 2- to 10-fold over control. In contrast, the PXR reporter gene assay showed less variation. For example, rifampin (2 μM) activated the PXR reporter gene 21- to 25-fold over control in three separate experiments.
The present study shows that the PXR reporter gene assay and primary culture of human hepatocytes produced similar but not identical CYP3A4 induction results. Clotrimazole, phenobarbital, rifampin, and sulfinpyrazone highly activate PXR and increased CYP3A4 activity; carbamazepine, dexamethasone, dexamethasone-t-butylacetate, phenytoin, sulfindimidine, and taxol weakly activated PXR and induced CYP3A4 activity; and methotrexate and probenecid showed no marked activation in either system. Ritonavir and troleandomycin showed marked PXR activation but no increase (in case of troleandomycin) or a significant decrease (in case of ritonavir) in microsomal CYP3A4 activity. The results generated from the correlation analysis support this concept. The correlation coefficient r was significantly increased from 0.529 to 0.864 (P < 0.001) in the absence of ritonavir and troleandomycin. In addition, a modest correlation was observed between PXR activation and induction of CYP3A4 mRNA levels.
Troleandomycin potently activated PXR in the reporter gene assay but did not increase the respective CYP3A4 activity and mRNA level. It is possible that the PXR reporter gene (luciferase) assay is more sensitive than the conventional CYP3A4 induction assay using human hepatocytes. Troleandomycin at the concentrations of 20 μM or less in this study may not be high enough to induce CYP3A4 in hepatocytes (Pichard et al., 1990; Ledirac et al., 2000). In addition, as a potent quasi-irreversible inhibitor of CYP3A4 (Chan et al., 1998; Greenblatt et al., 1998; Hamaoka et al., 2001), troleandomycin inhibits CYP3A4 activity and accelerates CYP3A4 enzyme inactivation.
Sulfadimidine has been shown to induce CYP3A4 activity at 50 μM (Pichard et al., 1990), whereas the maximal induction of CYP3A4 by dexamethasone occurs at near millimolar concentrations (LeCluyse et al., 2000). Taxol did not induce CYP3A4 in the present study, which differs from previous reports (Kostrubsky et al., 1998, 1999) in which Williams medium instead of modified Chee's medium was used, and testosterone 6β-hydroxylase activity was determined by directly adding testosterone to the medium and incubating for 30 min, instead of preparing microsomes from cultured hepatocytes and incubating for 10 min under different culture and anlytical conditions. In addition, individual variability may partially explain the difference observed.
There are several lines of evidence to support the working hypothesis that ritonavir is a CYP3A4 inducer. It highly increased PXR reporter gene activity, suggesting its intrinsic CYP3A4 induction potential. Correspondingly, the levels of CYP3A4 mRNA and protein were increased in the ritonavir-treated human hepatocytes. And one of six livers (H254) showed a 6-fold increase in the CYP3A4 activity by ritonavir at 2 μM. Ritonavir has been reported to accelerate the CYP3A4-mediated metabolism of dapsone, ethinyl estradiol, and methadone in healthy volunteers or in patients after 10 to 16 days administration (Gass et al., 1998; Ouellet et al., 1998; Geletko and Erickson, 2000). Furthermore, the primary metabolic route of ritonavir in humans is CYP3A4, and in vivo it exhibits concentration-dependent autoinduction (Hsu et al., 1997).
The observation that CYP3A4 activity from ritonavir-treated human hepatocytes was much lower than the control, and even became undetectable at 20 μM, could be explained by its potent inhibition, in particular by reversible and irreversible mechanisms of CYP3A4 inactivation (Kumar et al., 1996, 1999; Eagling et al., 1997;Koudriakova et al., 1998). In vitro, it inhibits CYP3A4-mediated metabolism of nifedipine (IC50 = 70 nM), ABT-378 (Ki = 13 nM), and testosterone (Ki = 19 nM). In vivo, it significantly decreases the metabolism of CYP3A4 substrates including clarithromycin, ketoconazole, indinavir, and saquinavir (Hsu et al., 1998a).
Notably, a particular good relationship between CYP3A4 mRNA levels (and activity) and PXR activation was observed in this study for compounds that elicited changes between 1- and 5-fold, especially when outliers, such as ritonavir, troleandomycin, and taxol, are removed from consideration for reasons outlined above. However, further activation of PXR by a number of compounds did not lead necessarily to greater CYP3A4 expression in primary hepatocytes. For example, compounds that generated between 5- and 25-fold increases in PXR activation caused only modest increases between 3- and 4-fold in CYP3A4 mRNA and activity. These results suggest that there may be molecular or biochemical limitations to the amount of CYP3A4 gene expression elicited from further drug-induced activation of nuclear receptors or negative feedback regulation by CYP3A4 itself (Schuetz and Strom, 2001). The clinical implications of this phenomenon, if real, are not known at this point but may have important repercussions on the extrapolation of data derived from transfection assays and the prediction of enzyme induction in vivo.
In cultured human hepatocytes and in vivo, PXR represents the main signaling pathway for CYP3A4 induction. However, other transcriptional factors such as CAR may also participate in the regulation of CYP3A4. In addition to activating the CYP2B phenobarbital responsive enhancer module, CAR could directly transactivate the CYP3A4 xenobiotic responsive element (ER6/DR3), which serves as a PXR/retinoid X receptor binding site (Sueyoshi et al., 1999; Moore et al., 2000; Xie et al., 2000). Phenobarbital probably induces CYP3A4 through more than one pathway. At high concentrations, phenobarbital directly activated human PXR as presented in this study and reported from other laboratories (Jones et al., 2000; Moore et al., 2000). In addition, phenobarbital and phenobarbital-like compounds were demonstrated to induce CYP3A4 gene through CAR-mediated signaling pathway (Sueyoshi et al., 1999; Xie et al., 2000). Phenobarbital, but not dexamethasone, was reported to induce CYP3A11 in PXR gene knockout mice (Staudinger et al., 2001).
In summary, results from the PXR reporter gene assay and primary culture of human hepatocytes were generally in agreement. A discrepancy between PXR activation and CYP3A4 activity was observed for ritonavir and troleandomycin due to irreversible enzyme inhibition and concomitant induction. Also, the PXR reporter gene assay cannot reveal the ability to induce CYP3A4 via the CAR pathway (e.g., phenobarbital). On the other hand, the enzyme-inducing potential of ritonavir in cultured human hepatocytes would, in most cases, be overlooked unless mRNA and/or protein levels were determined in addition to measurement of CYP3A4 activity. These results show that both the PXR reporter gene assay and primary cultures of human hepatocytes can be used in a complementary fashion to determine the overall induction potential of new drugs under development.
Acknowledgments
We thank Dr. Eric Solon and Leifei Wang for scanning and analysis of Western blots.
Footnotes
- Abbreviations used are::
- PXR
- pregnane X receptor
- DMSO
- dimethylsulfoxide
- bDNA
- branched DNA
- CAR
- constitutive androstane receptor
- ABT-378
- lopinavir
- Received November 7, 2001.
- Accepted March 22, 2002.
- The American Society for Pharmacology and Experimental Therapeutics