Abstract
Glucuronidation is a major detoxification pathway of drugs and xenobiotics that are catalyzed by the UDP-glucuronosyltransferase (UGT) superfamily. Determination of the protein levels of the individual UGT isoforms in human tissues is required for the successful extrapolation of in vitro metabolic data to in vivo clearance. Most previous studies evaluating UGT isoform expression were limited to the mRNA level because of the high degree of amino acid sequence homology between UGT isoforms that has hampered the availability of isoform-specific antibodies. In this study, we generated a peptide-specific monoclonal antibody against human UGT1A9. We demonstrated that this antibody does not cross-react with the other UGT1A isoforms including UGT1A7, UGT1A8, and UGT1A10 and shows a high degree of amino acid sequence similarity with UGT1A9. Using this antibody, we found that UGT1A9 protein is expressed in the kidney and the liver but not in the jejunum or the ileum, consistent with previous reports of mRNA expression. In a panel of 20 individual human livers, the UGT1A9 protein levels exhibited 9-fold variability. It is noteworthy that the relative UGT1A9 protein levels were not correlated with the UGT1A9 mRNA level (r = −0.13), like other UGT isoforms reported previously, suggesting the importance of evaluating UGT isoform expression at protein levels. In conclusion, we generated a specific monoclonal antibody against UGT1A9 and evaluated the distribution and relative expression levels of the UGT1A9 protein in human tissues. This antibody may serve as a useful tool for further studies of UGT1A9 to evaluate its physiological, pharmacological, and toxicological roles in human tissues.
Introduction
UDP-glucuronosyltransferases (UGTs) are a family of phase II drug-metabolizing enzymes that play key roles in the metabolism of endogenous and exogenous compounds. UGTs mediate the transfer of glucuronic acid from UDP-glucuronic acid to hydrophobic compounds, facilitating their elimination via bile and urine. Whereas glucuronidation usually inactivates biologically active molecules, there are exceptions, such as morphine and retinoic acids, which are converted to pharmacologically active glucuronides (Shimomura et al., 1971; Formelli et al., 1996).
Human UGTs are classified by evolutionary divergence into three subfamilies, including UGT1A, UGT2A, and UGT2B (Mackenzie et al., 2005). The human UGT1A gene cluster is located on chromosome 2q37 and contains multiple unique first exons, as well as the conserved exons 2 to 5, which can give rise to nine kinds of functional UGT1A isoforms, including UGT1A1, UGT1A3, UGT1A4, UGT1A5, UGT1A6, UGT1A7, UGT1A8, UGT1A9, and UGT1A10 (Gong et al., 2001). Among them, UGT1A1, UGT1A3, UGT1A4, UGT1A6, and UGT1A9 are expressed in liver, whereas UGT1A7, UGT1A8, and UGT1A10 are predominantly expressed in the gastrointestinal tract (Strassburg et al., 1997, 2000). In these tissues, UGT enzymes are induced by various endogenous and exogenous compounds (Sutherland et al., 1993; Mackenzie et al., 2003). Genetic polymorphisms have been reported for most human UGT genes (Guillemette, 2003). These features would be the possible reasons for interindividual variability of expression level and enzymatic activity, which may be associated with the interindividual variability of drug efficacy and toxicity.
Variability of UGT expression can be evaluated by measuring enzyme activity in vitro using specific substrates. However, substrates that are specifically metabolized by a single UGT isoform are limited because of the broad and overlapping substrate specificities of UGTs. Inappropriate selection of substrates may lead to misevaluation. An alternative approach is to measure mRNA levels of individual UGT isoforms. Indeed, earlier studies evaluated the interindividual difference in the expression or tissue distribution of UGT isoforms at the mRNA level (Strassburg et al., 2000; Izukawa et al., 2009; Ohno and Nakajin, 2009). However, we and other research groups reported that certain individual UGT mRNA levels correlate poorly with their respective protein levels (Izukawa et al., 2009; Ohtsuki et al., 2012). Therefore, it should be noted that the mRNA levels might not necessarily reflect the actual UGT protein levels. Immunochemical techniques are the most conventional approaches for the assessment of protein levels. However, this approach is limited by the specificity of available antibodies. At present, specific antibodies for human UGT isoforms are only commercially available for UGT1A1 and UGT1A6. Antibodies against UGT1A4 and UGT1A9 are also available for purchase, but their specificity is not guaranteed. Because UGT1A9 exhibits as high as 88 to 94% amino acid homology with UGT1A7, UGT1A8, and UGT1A10, it is a great challenge to generate a specific antibody against UGT1A9. Indeed, the antibody against UGT1A9 prepared by Girard et al. (2004) exhibits cross-reactivity against UGT1A7, UGT1A8, and UGT1A10. In this study, we generated a highly specific monoclonal antibody against human UGT1A9 to evaluate the variability of UGT1A9 protein levels in a panel of human liver samples by Western blot analysis.
Materials and Methods
Materials.
Recombinant human UGT1A1, UGT1A3, UGT1A4, UGT1A6, UGT1A7, UGT1A8, UGT1A9, and UGT1A10 expressed in baculovirus-infected insect cells (Supersomes) and human liver microsomes (a pooled sample, n = 50) were purchased from BD Gentest (Woburn, MA). Human kidney microsomes (a pooled sample, n = 6, as well as three individual donors), human jejunum microsomes (a pooled sample, n = 10), and human ileum microsomes (a pooled sample, n = 4) were purchased from Tissue Transformation Technologies (Edison, NJ). Human liver samples from 14 individual donors were supplied by the National Disease Research Interchange (Philadelphia, PA) through the Human and Animal Bridging Research Organization (Chiba, Japan), and those from 6 Japanese donors were obtained from autopsy materials that were discarded after pathological investigation (Izukawa et al., 2009). Microsomes were prepared as described previously (Tabata et al., 2004). The use of the human livers was approved by the ethics committees of Kanazawa University (Kanazawa, Japan) and Iwate Medical University (Morioka, Japan). A human embryonic kidney-derived cell line (HEK293) stably expressing UGT1A9 was established previously in our laboratory (Fujiwara et al., 2007). A human hepatocellular carcinoma cell line (HepG2), an immortalized human kidney tubular epithelial cell line (HK-2), and a human breast adenocarcinoma cell line (MCF-7) were obtained from American Type Culture Collection (Manassas, VA) and cultured as described previously (Nakamura et al., 2008). Endoglycosidase H (Endo H) and peptide:N-glycosidase F (PNGase F) were purchased from New England Biolabs (Ipswich, MA). A goat anti-human hepatocyte nuclear factor (HNF) 1α (C-19) polyclonal antibody and mouse anti-β-actin monoclonal antibody (C-14) were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). All the other reagents were of the highest grade commercially available.
Preparation of Monoclonal Antibody against UGT1A9.
The selection of antigenic peptide, peptide synthesis, and keyhole limpet hemocyanin conjugation was performed by Biogate (Gifu, Japan). Hydrophilicity, secondary structure, surface probability, and antigenicity were considered in the designation of the antigenic peptide sequence as follows. The hydrophilicity was evaluated by the method of Hopp and Woods (1981). The secondary structure was evaluated by the method of Chou and Fasman (1974) and the method of Robson (Garnier et al., 1978) using GENETYX-MAC software (Software Development, Tokyo, Japan). The surface probability was evaluated by the method of Emini et al. (1985). Antigenicity was evaluated by the method of Welling et al. (1985) and the method of Parker et al. (1986) using original software. The designed peptide sequence was subjected to BLASTP search (http://www.ncbi.nlm.nih.gov/blast/) to screen its homology with known protein sequences. Based on these evaluations, the sequence, DREFKAFAHAQWKAQVRSIYSLLMGSYNDIFD, which corresponds to the residues 87 to 118 of UGT1A9, was raised as a candidate peptide. At the N terminus of the synthesized peptide, a cysteine residue was added to facilitate conjugation to the carrier protein, keyhole limpet hemocyanin. The mouse monoclonal antibody against the peptide was prepared by CLEA Japan (Tokyo, Japan) using a standard protocol. The hybridomas producing the antibodies were screened by enzyme-linked immunosorbent assay with the synthesized peptide. Reactivity and specificity of antibody clones were evaluated by Western blotting as described below. A clone that specifically reacted with UGT1A9 was expanded by intraperitoneal injection into mineral oil-primed mice. Monoclonal antibodies from mouse ascitic fluids were partially purified by precipitation with 33% ammonium sulfate.
SDS-PAGE and Western Blot Analysis.
For the analysis of UGT1A9, UGT Supersomes (2.5 μg), total cell homogenates from HEK293 cells expressing UGT1A9 (40 μg), or microsomes from human tissues (15 μg), mouse or rat liver, and human cell lines (30 μg) were separated by 10% SDS-PAGE and transferred to Protran nitrocellulose membranes (Whatman GmbH, Dassel, Germany). The quantity of protein loaded onto gels was decided to be in the range showing linearity. In some cases, human liver microsomes or recombinant UGT1A9 proteins were treated with Endo H, which cleaves the bond between two N-acetylglucosamines directly proximal to the asparagine residue, or PNGase F, which cleaves the bond between asparagine and the N-acetylglucosamine residue. The enzyme sources were adjusted to a 2 mg/ml protein concentration with a denaturing buffer containing a final concentration of 0.5% SDS and 40 mM dithiothreitol and subsequently were denatured at 95°C for 10 min. An aliquot [2.5 μg of UGT1A9 Supersomes, 40 μg of UGT1A9 expressed in HEK293 cells, and 30 μg of human liver microsomes (HLM)] was incubated with 250 U of Endo H in 50 mM sodium citrate buffer (pH 5.5) or 500 U of PNGase F in 50 mM sodium phosphate buffer (pH 7.5) containing 1% NP-40 at 37°C for 1 h.
For the analysis of HNF1α, 50 μg of human liver homogenates were separated by 7.5% SDS-PAGE and transferred to a polyvinylidene difluoride Immobilon-P membrane (Millipore Corporation, Billerica, MA). For the analysis of β-actin, 10 μg of human liver microsomes or homogenates were separated by 7.5% SDS-PAGE and transferred to polyvinylidene difluoride membranes.
After incubation in Odyssey blocking buffer (LI-COR Biosciences, Lincoln, NE) for 1 h, the membranes were probed with either 1:500 diluted anti-UGT1A9 antibody, 1:200 diluted anti-HNF1α antibody, or 1:200 diluted anti-β-actin antibody for 3 h followed by incubation with the corresponding fluorescent dye-conjugated secondary antibodies. The UGT1A9 or HNF1α protein levels in individual human liver samples were normalized with β-actin protein levels. The band densities were quantified with the Odyssey Infrared Imaging system (LI-COR Biosciences).
Propofol O-Glucuronosyltransferase Activity.
The activity was determined as described previously (Fujiwara et al., 2007) with a substrate concentration of 500 μM.
Statistical Analyses.
Correlation analyses were performed by the Pearson product-moment method. Differences between groups were determined by analysis of variance followed by the Tukey multiple comparison test. p < 0.05 was considered statistically significant.
Results
Selection of a Peptide Antigen to Generate UGT1A9 Antibody.
Initially, preparation of a mouse monoclonal antibody against UGT1A9 was attempted using a histidine-tagged full-length UGT1A9 protein as an antigen. However, all of the resulting antibody clones (30 clones) cross-reacted with other UGT1A isoforms (data not shown). The full-length amino acid sequence of UGT1A9 exhibits homology with UGT1A7, UGT1A8, and UGT1A10 in excess of 88% (Table 1). Therefore, we next sought to prepare the monoclonal antibody using a UGT1A9 peptide as an antigen. Although peptides ranging from 10 to 20 amino acid residues in length are generally used for antigens, we used a relatively longer peptide, expecting that it could recognize three-dimensional structure. Residues 87 to 118 of UGT1A9 (32 amino acids) were selected as an antigen; this peptide sequence is contained within the longer peptide antigen (82 amino acids, residues 61–142) used by Girard et al. (2004) (Table 1). The amino acid homology of the two different antigenic peptides with the corresponding residues of UGT1A7, UGT1A8, and UGT1A10 was 59 to 63% (32-residue peptide) versus 80% (82-residue peptide). Thus, the peptide comprising the 87 to 118 amino acid residues of UGT1A9 was used as an antigen.
Sequence alignment around the candidate peptide of UGT1A9 as an antigen with the other UGT1As
Multiple sequence alignment was performed using the ClustalW2 program (http://www.ebi.ac.uk/Tools/msa/clustalw2/) for amino acid residues 61 to 142 of UGT1A9, which were used as an antigen by Girard et al. (2004). Amino acids that are identical with those of UGT1A9 are in bold type. The peptide comprising amino acid residues 87 to 118 that was used as antigen in this study is in italic type.
Specificity of the Prepared Antibody against UGT1A9.
The specificity of the 40 candidate antibody clones was evaluated by Western blot analysis using a panel of recombinant human UGT1A isoforms. Of the 40 clones, 5 reacted with UGT1A9 without cross-reacting with the other UGT1A isoforms (data not shown), from which the clone exhibiting the highest reactivity was selected for expansion and antibody production. The specificity of the purified antibody was then confirmed (Fig. 1A). Next, we investigated whether the antibody reacted with UGTs in human, mouse, and rat liver microsomes (Fig. 1B). The mouse Ugt1a9, Ugt1a7c, and Ugt1a10 exhibit 77 to 78% amino acid sequence homology with human UGT1A9 (Table 2). In rat, the Ugt1a9 gene is a pseudogene, whereas Ugt1a7, Ugt1a8, and Ugt1a10, which are functional proteins, exhibit 76 to 79% amino acid homology with human UGT1A9. As shown in Fig. 1B, a clear single band was observed only with human liver microsomes, suggesting that the antibody does not react with any Ugt in mouse and rat livers.
Western blot analyses using the monoclonal antibody against human UGT1A9. A, recombinant UGT1As (10 μg) expressed in baculovirus-infected insect cells (Supersomes). B, pooled microsomes from human, mouse, or rat liver (30 μg). C, Endo H-treated (left) or PNGase F-treated (right) (+) and nontreated (−) UGT1A9 Supersomes (2.5 μg), recombinant UGT1A9 stably expressed in HEK293 (40 μg), and HLM (30 μg). D, microsomes from human liver, kidney, jejunum, and ileum (15 μg). E, microsomes from human liver and human cell lines (30 μg). In C, the arrowhead and asterisk represent UGT1A9 and the nonspecific band, respectively. MLM, mouse liver microsomes; RLM, rat liver microsomes; HKM, human kidney microsomes; HJM, human jejunum microsomes; HIM, human ileum microsomes; M, marker.
Sequence alignment of the candidate peptide of human UGT1A9 as an antigen with highly similar rodent Ugt1a isoforms
Multiple sequence alignment was performed using the ClustalW2 program (http://www.ebi.ac.uk/Tools/msa/clustalw2/) for amino acid residues 87 to 118 and those of the corresponding region of rodent Ugt1a isoforms. Amino acids that are identical with those of UGT1A9 are in bold type.
Reactivity of the Antibody toward Glycosylated or Deglycosylated UGT1A9.
We previously reported that UGT1A9 is glycosylated at three asparagine residues at position 71, 292, and 344 (Nakajima et al., 2010). Three bands observed in UGT1A9 Supersomes (Fig. 1A) would represent differently glycosylated species of UGT1A9, because none of them were observed in the other UGT1A Supersomes. We investigated whether the antibody could recognize both glycosylated and deglycosylated or unglycosylated UGT1A9. When the UGT1A9 Supersomes were treated with Endo H, the upper two bands observed in the nontreated sample disappeared, and a band with higher mobility was observed. The density of the faster migrating band appeared higher than that of the sum of the upper two bands (Fig. 1C, left). The fastest migrating band in the nontreated sample might be the glycosylated form that is tolerable to Endo H or other post-translationally modified form. The recombinant UGT1A9 stably expressed in HEK293 cells also showed three bands (Fig. 1C, left), but the upper two bands would be nonspecific bands because they were observed in homogenates from mock HEK293 cells too (data not shown). The difference in the band patterns between UGT1A9 Supersomes and UGT1A9 in HEK293 cells might reflect the differences in the extent of glycosylation and/or size of the glycan in insect or mammalian cells. When the recombinant UGT1A9 in HEK293 cells was treated with Endo H, the fastest migrating band was clearly shifted (Fig. 1C, left). The band density was higher than that in the nontreated sample. As for HLM, the mobility of UGT1A9 was similar to that of UGT1A9 expressed in HEK293 cells, and UGT1A9 in HLM appeared to show some tolerance to Endo H. Next, we used PNGase F, which can cleave Endo H-resistant N-glycans (probably N-glycans from which two mannose subunits are removed by Golgi α-mannosidase II). By the treatment of HLM with PNGase F, only a band with faster mobility was observed, indicating that the upper band observed in Endo H-treated HLM would be the Endo H-resistant glycosylated UGT1A9 (Fig. 1C, right). In the cases of UGT1A9 Supersomes and UGT1A9 in HEK293 cells, the results with PNGase F treatment were the same as those with Endo H treatment. It is interesting that the deglycosylated UGT1A9 in UGT1A9 Supersomes and UGT1A9 in HEK293 cells and HLM differently migrated, although it is still unclear whether other post-translational modifications such as phosphorylation may be involved. Taken together, these results suggest that the antibody can recognize UGT1A9 regardless of glycosylation status, although the reactivity seems to be enhanced for unglycosylated UGT1A9.
UGT1A9 Protein Expression in Human Tissues.
Previous studies reported that UGT1A9 mRNA is predominantly expressed in the liver and the kidney, and, to a much lesser extent, in adrenal, colon, small intestine, esophagus, testis, and bladder tissues (Nakamura et al., 2008; Ohno and Nakajin, 2009). We obtained microsomes from human liver, kidney, jejunum, and ileum, which we subjected to Western blot analysis. As shown in Fig. 1D, high expression of UGT1A9 protein was detected in the kidney, followed by the liver, but negligible expression was observed in the jejunum and the ileum, consistent with the previously reported mRNA expression profiles. To investigate whether UGT1A9 expression is generally higher in the kidney than in the liver, liver and kidney microsomes obtained from three individuals were subjected to Western blot analysis. We found that the relative UGT1A9 protein levels are approximately 3 to 16 times higher in the kidney than in the liver.
UGT1A9 Protein Expression in Human Cell Lines.
We previously reported that UGT1A9 is detectable at the mRNA level in human cell lines, including HepG2, HK-2, and MCF-7 (Nakamura et al., 2008), for which reproducibility was confirmed by quantitative real-time reverse transcriptase-polymerase chain reaction (data not shown). However, to date, there has been no reported quantitation of UGT1A9 at the protein level in human cell lines using an isoform-specific antibody. We investigated whether UGT1A9 protein could be detected in HepG2, HK-2, and MCF-7 cell lines using the antibody that we generated. When the microsomes from these cell lines were subjected to Western blot analysis, no band was observed, which is probably attributed to low UGT1A9 expression levels in these cells (Fig. 1E).
Normalized Activities of UGT1A9 in Recombinant Systems and Human Tissue Microsomes.
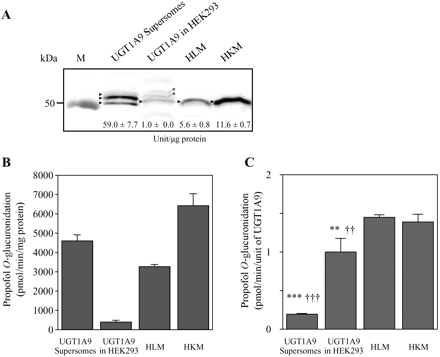
The prepared specific antibody against UGT1A9 enabled us to determine the normalized activities per unit of UGT1A9 in recombinant systems and human tissue microsomes. We determined the relative expression levels of UGT1A9 protein in UGT1A9 Supersomes and recombinant UGT1A9 stably expressed in HEK293 cells, HLM, and human kidney microsomes as 59 ± 7.7, 1.0 ± 0.0, 5.6 ± 0.8, and 11.6 ± 0.7 units/μg protein, respectively (Fig. 2A). The propofol O-glucuronidation activities in these enzyme sources were 4598 ± 309, 400 ± 98, 3275 ± 99, and 6418 ± 635 pmol · min−1 · mg protein−1, respectively (Fig. 2B). Accordingly, it was demonstrated that the normalized activities per unit of UGT1A9 in UGT1A9 Supersomes and recombinant UGT1A9 in HEK293 cells were 7- and 1.4-fold, respectively, lower than those in human tissue microsomes (Fig. 2C).
UGT1A9 protein expression and propofol O-glucuronidation in recombinant UGT1A9 expression systems and human tissue microsomes. A, Western blot analysis of UGT1A9 protein in UGT1A9 Supersomes (2.5 μg) and recombinant UGT1A9 stably expressed in HEK293 (40 μg), HLM (15 μg), and human kidney microsomes (HKM) (15 μg). The values shown in the membrane indicate the expression level of UGT1A9 expressed as units of UGT1A9 per microgram of protein. Data are the means ± S.D. of quadruplicate determinations. The arrowhead and asterisk represent UGT1A9 and the nonspecific band, respectively. B and C, propofol O-glucuronidation activity in these enzyme sources expressed as picomoles per minute per milligram of protein (B) and picomoles per minute per unit of UGT1A9 protein (C). Columns represent the means ± S.D. of triplicate determinations. **, p < 0.01; ***, p < 0.001, compared with HLM; ††, p < 0.01; †††, p < 0.001, compared with HKM. M, marker.
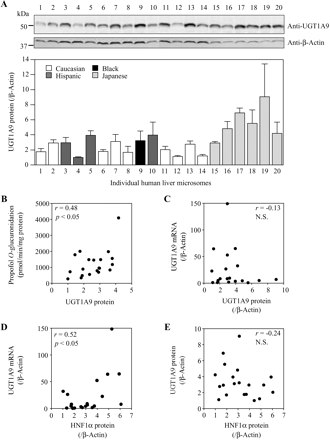
Expression Levels of UGT1A9 Protein in Individual Human Livers and the Correlation of Protein with mRNA Levels and Enzymatic Activity.
We assessed the relative expression levels of UGT1A9 protein in a panel of 20 human liver microsomes and found a 9-fold interindividual variability (Fig. 3A). The UGT1A9 protein level normalized with β-actin seems to be higher in Japanese than in other ethnic groups. However, it may be due to a lower β-actin level in Japanese than in the other ethnic groups. To draw a definitive conclusion for the ethnic difference, analysis with a larger sample size is necessary. The UGT1A9 protein levels were moderately correlated (r = 0.48, p < 0.05) with propofol O-glucuronosyltransferase activity (Fig. 3B). However, the UGT1A9 protein levels were not correlated (r = −0.13) with the UGT1A9 mRNA levels that were determined in our previous study (Fig. 3C), exhibiting a 150-fold variability (Izukawa et al., 2009). Previous studies (Aueviriyavit et al., 2007; Ramírez et al., 2008) reported that HNF1α and HNF4α mRNA levels were strongly correlated with UGT1A9 mRNA levels in a panel of human livers, which is consistent with the finding that HNF1α and HNF4α contribute to the regulation of UGT1A9 (Barbier et al., 2005; Gardner-Stephen and Mackenzie, 2007). However, the HNF protein levels rather than mRNA level should be considered. Thus, we investigated whether the HNF1α and HNF4α protein levels are correlated with the UGT1A9 mRNA or protein levels. We found that the HNF1α protein levels were significantly correlated with the UGT1A9 mRNA levels (r = 0.52, p < 0.05) but not with the UGT1A9 protein levels (r = −0.24, not significant) (Fig. 3, D and E). These data suggest that whereas HNF1α may regulate UGT1A9 at the transcriptional level, UGT1A9 protein levels are being regulated at the post-transcriptional level. Unfortunately, we could not assess the HNF4α protein levels because of an insufficient dynamic range of the antibody used (data not shown).
Interindividual variability of UGT1A9 protein levels in human liver and its correlation with enzyme activity, UGT1A9 mRNA, or HNF1α protein levels. A, expression levels of UGT1A9 protein in 20 human liver microsomes were determined by Western blot analysis. Data are the means ± S.D. of triplicate determinations. Relationships between UGT1A9 protein levels and propofol O-glucuronosyltransferase activities (B), between UGT1A9 protein levels and UGT1A9 mRNA levels (C), between HNF1α protein levels and UGT1A9 mRNA levels (D), and between HNF1α protein levels and UGT1A9 protein levels (E) were analyzed. The propofol O-glucuronosyltransferase activity was measured at a substrate concentration of 500 μM. The UGT1A9 mRNA levels were normalized to β-actin mRNA levels. The UGT1A9 and HNF1α protein levels were normalized to β-actin protein levels. The values represent the levels relative to that of the lowest sample. Each data point is the mean of duplicate experiments except UGT1A9 protein level. N.S., not significant.
Discussion
Human UGT1A9 is one of the physiologically and pharmacologically important UGT1A isoforms, metabolizing a wide spectrum of substrates including bulky phenols, dietary constituents, steroids, and fatty acids, as well as currently prescribed drugs including anticancer agents, fibrates, nonsteroidal anti-inflammatory drugs and antiarrhythmic agents (Ritter, 2000). Human UGT1A9 mRNA was found to be predominantly expressed in liver and kidney and to a lesser extent in esophagus, small intestine, colon, adrenal gland, and bladder (Nakamura et al., 2008; Ohno and Nakajin, 2009). However, further information regarding UGT1A9 protein has been limited by the lack of a specific antibody against UGT1A9. Although there were previous attempts to generate specific antibodies against UGT1A9 (Girard et al., 2004; Ikushiro et al., 2006), the antibodies recognized other UGT1A isoforms such as UGT1A6 (Ikushiro et al., 2006), UGT1A7, UGT1A8, and UGT1A10 (Girard et al., 2004). Furthermore, when we evaluated the specificity of commercially available antibodies against UGT1A9 (Abcam, Cambridge, UK; Abnova, Taipei, Taiwan), we observed that these antibodies cross-reacted with other UGT1A isoforms (S. Oda and M. Nakajima, unpublished data). This background prompted us to prepare a specific antibody against human UGT1A9. We used a peptide containing the amino acid residues 87 to 118 of UGT1A9 as an antigen. Although this peptide is relatively longer than those generally used as antigens, it is shorter than the peptide antigen used by Girard et al. (2004) that contained UGT1A9 amino acid residues 61 to 142. Here, we report the first successful generation of an antibody that specifically recognizes UGT1A9.
Upon Western blotting using the prepared antibody, we confirmed that there was no aggregated UGT1A9 at the interface between the upper and lower gels or the bottom of the wells in any enzyme source (data not shown). The expression level of UGT1A9 protein was found to be highest in the kidney, followed by the liver, but was negligible in jejunum and ileum (Fig. 1D), consistent with previously reported mRNA data (Nakamura et al., 2008; Ohno and Nakajin, 2009). Although the kidney plays a role in the excretion of polar xenobiotics and metabolites, increasing evidence reveals that the kidney significantly contributes to metabolic clearance of therapeutic drugs, such as nonsteroidal anti-inflammatory drugs, propofol, and mycophenolic acid, and to the maintenance of renal homeostasis through inactivating mediators, such as prostaglandins, leukotrienes, epoxyeicosatrienoic acids, and hydroxyeicosatetraenoic acids (Knights and Miners, 2010). Because these drugs or endobiotics are known to be substrates of UGT1A9 (Knights and Miners, 2010), it has been speculated that UGT1A9 would contribute to their clearance. There has been only one report of the immunohistochemical detection of UGT1A (Gaganis et al., 2007), although the precise isoforms detected remain unknown. The present study supports the role of UGT1A9 in the kidney, as indicated by the substantial expression of UGT1A9 protein detected by Western blotting. The antibody that we prepared will be useful for future immunohistochemical studies of UGT1A9.
An interesting finding using the prepared antibody was that the normalized activities of UGT1A9 in recombinant systems were unambiguously lower than those in human tissue microsomes (Fig. 2). This might be attributable to the differences in membrane circumstance including lipid components and/or post-translational modification between recombinant systems and human tissue microsomes. Another possible explanation is the presence of other UGT isoforms in human tissue microsomes. Previous studies (Fujiwara et al., 2007, 2010) demonstrated that the coexpression of another UGT isoform increases the UGT1A9-catalyzed propofol O-glucuronidation in HEK293 cells. Apart from these reasons, it is notable that the activities of recombinant UGTs do not directly and quantitatively mirror the actual UGT activities in human tissues. In this regard, the relative activity factor approach (Crespi and Miller, 1999), which uses the ratio of activity of tissue microsomes and recombinant enzymes, would be useful to estimate the contributions of individual UGTs to a given metabolic pathway in tissue microsomes, as recent studies reported (Kato et al., 2012; Zhu et al., 2012).
We found moderate interindividual variability (9-fold) of hepatic UGT1A9 expression at the protein level (Fig. 3). Several studies have sought to uncover the underlying mechanisms of the variability in the UGT1A9 expression, with particular focus on cis- or trans-acting factors. As cis-acting factors, genetic polymorphisms can be raised. The −275 T>A and −2152 C>T alleles, which are linked to each other, have been shown to be associated with higher hepatic UGT1A9 protein expression and increased rates of propofol and mycophenolic acid glucuronidation (Girard et al., 2004). In addition, homozygotes for the intronic single nucleotide polymorphism at position IVS1 +399C>T have been shown to exhibit higher (1.3-fold) hepatic UGT1A9 protein levels (Girard et al., 2006). In these studies, UGT1A9 protein was assessed using the UGT1A7–10 antibody. The fact that UGT1A7, UGT1A8, and UGT1A10 are not expressed in liver made such studies possible. In contrast, our antibody would be applicable for the evaluation of the effects of this single nucleotide polymorphism on UGT1A9 expression in extrahepatic tissues expressing UGT1A9 and the closely related isoforms UGT1A7, UGT1A8, and UGT1A10, such as kidney and adrenal tissues (Ohno and Nakajin, 2009). As trans-acting factors, transcription factors can be raised. It has been reported that HNF1α and HNF4α positively regulate the expression of UGT1A9 (Barbier et al., 2005; Gardner-Stephen and Mackenzie, 2007). A significant positive correlation between these factors and UGT1A9 at the mRNA level in human livers has been reported (Aueviriyavit et al., 2007; Ramírez et al., 2008). Beyond these reports, we demonstrated that HNF1α protein levels are significantly correlated with UGT1A9 mRNA levels (Fig. 3D). However, HNF1α protein levels were not correlated with UGT1A9 protein levels (Fig. 3E), because we did not detect any correlation between the UGT1A9 mRNA and protein levels (Fig. 3C). Lack of correlation between the mRNA and protein levels has also been observed with other UGTs, such as UGT1A4, UGT1A6, and UGT2B7 (Izukawa et al., 2009). Therefore, it is reasonable to speculate that post-transcriptional and/or post-translational regulation plays a role in UGT protein levels. MicroRNAs (miRNAs) have recently received considerable attention as a critical factor of post-transcriptional regulation. We have reported that some cytochrome P450 isoforms and transcriptional factors, such as pregnane X receptor, vitamin D receptor, and HNF4α are regulated by miRNAs (Nakajima and Yokoi, 2011), implying that miRNAs have a role in clearance of drugs and endobiotics. It would be of interest to investigate whether miRNAs may be involved in the regulation of UGTs. In general, mammalian miRNAs bind to the 3′-untranslated region of the target mRNA to cause translational repression or mRNA degradation. Because the 3′-untranslated sequences of UGT1As are common, it is possible that UGT1As may be commonly regulated by the same miRNA.
The UGT1A9 protein levels in a panel of 20 individual human livers were moderately (r = 0.48, p < 0.05) correlated with propofol glucuronidation (Fig. 2B). The moderate correlation was consistent (r = 0.5, p < 0.0001, n = 48) with the results reported by Girard et al. (2004). Because we previously demonstrated that UGT enzyme activity could be modulated through formation of heterodimers with other UGT isoform (Fujiwara et al., 2007), such modulation might account for the moderate correlation.
In summary, we generated a specific monoclonal antibody against human UGT1A9. By Western blot analysis using this antibody, we found that human UGT1A9 protein is highly expressed in the kidney and the liver but not in the jejunum or the ileum, supporting previous findings at mRNA levels. This antibody can be used to assess tissue distribution and interindividual variability of UGT1A9 protein expression, and such evaluation may promote the understanding of the physiological, pharmacological, and toxicological role of UGT1A9.
Authorship Contributions
Participated in research design: Oda, Nakajima, Hatakeyama, Fukami, and Yokoi.
Conducted experiments: Oda.
Contributed new reagents or analytic tools: Hatakeyama.
Performed data analysis: Oda.
Wrote or contributed to the writing of the manuscript: Oda, Nakajima, and Yokoi.
Acknowledgments
We acknowledge Drs. Yasuhiro Aoki and Masataka Takamiya at Iwate Medical University for supplying human livers.
Footnotes
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
ABBREVIATIONS:
- UGT
- UDP-glucuronosyltransferase
- HEK
- human embryonic kidney
- Endo H
- endoglycosidase H
- PNGase F
- peptide:N-glycosidase F
- HNF
- hepatocyte nuclear factor
- PAGE
- polyacrylamide gel electrophoresis
- HLM
- human liver microsomes
- miRNA
- microRNA.
- Received March 8, 2012.
- Accepted May 22, 2012.
- Copyright © 2012 by The American Society for Pharmacology and Experimental Therapeutics