Visual Overview
Abstract
The aim of this study was to derive region-specific transporter expression data suitable for in vitro-to-in vivo extrapolation (IVIVE) within a physiologically based pharmacokinetic (PBPK) modeling framework. A meta-analysis was performed whereby literary sources reporting region-specific transporter expression obtained via absolute and relative quantification approaches were considered in healthy adult Caucasian individuals. Furthermore, intestinal total membrane protein yield was calculated to enable mechanistic IVIVE via absolute transporter abundances. Where required, authors were contacted for additional information. A refined database was constructed where samples were excluded based on quantification in, non-Caucasian subjects, disease tissue, subjects <18 years old, duplicated samples, non-total membrane matrix, pooled matrices, or cDNA. Demographic data were collected where available. The weighted and geometric mean, coefficient of variation, and between-study homogeneity was calculated in each of eight gut segments (duodenum, two jejunum, four ileum, and colon) for 16 transporters. Expression data were normalized to that in the proximal jejunum. From a total of 47 articles, the final database consisted of 2238 measurements for 16 transporters. The solute carrier peptide transporter 1 (PepT1) showed the highest jejunal abundance, while multidrug resistance-associated protein (MRP) 2 was the highest abundance ATP-binding cassette transporter. Transporters displaying significant region-specific expression included the ileal bile acid transporter, which showed 18-fold greater terminal ileum expression compared with the proximal jejunum, while MRP3, organic cation transporter type 1 (OCTN1), and OCT1 showed >2-fold higher expression in other regions compared with the proximal jejunum. This is the first systematic analysis incorporating absolute quantification methodology to determine region-specific intestinal transporter expression. It is expected to be beneficial for mechanistic transporter IVIVE in healthy adult Caucasians.
Significance Statement Given the burgeoning reports of absolute transporter abundances in the human intestine, the incorporation of such information into mechanistic IVIVE-PBPK models could offer a distinct advantage in facilitating the robust assessment of the impact of gut transporters on drug disposition. The systematic and formal assessment via a literature meta-analysis described herein, enables assignment of the regional-specific expression, absolute transporter abundances, interindividual variability, and other associated scaling factors to healthy Caucasian populations within PBPK models. The resulting values are available to incorporate into PBPK models, and offer a verifiable account describing intestinal transporter expression within PBPK models for persons wishing to utilize them. Furthermore, these data facilitate the development of appropriate IVIVE scaling strategies using absolute transporter abundances.
Introduction
Physiologically based pharmacokinetic (PBPK) models are increasingly being employed in making key decisions on the clinical progress of new chemical entities (NCEs) in drug development and regulatory spheres (Shebley et al., 2018). A growing assurance in the ability to predict the pharmacokinetics, pharmacodynamics, and drug-drug interactions of NCEs within virtual individuals built into PBPK models has been based on the continued dedication of allied academic, industrial, and regulatory institutions to develop robust physiologic parameters that are essential in facilitating the generation of relevant individuals within virtual populations. Alongside this, the development of mechanistic strategies that harness data generated from in vitro assays routinely performed to characterize the NCE pharmacokinetic liability via in vitro-to-in vivo extrapolation (IVIVE) strategies is critical in enabling insightful judgments regarding clinical progress being made. For several years now, the capacity to scale cytochrome P450 (P450) activities via recombinant in vitro systems and absolute protein expression has been demonstrated in the intestine (Gertz et al., 2010). Historically, the availability of protein standards employed within assays to quantify P450 absolute protein abundances in both in vitro systems and mammalian tissues has facilitated the development of such strategies. These approaches are underpinned by incorporating scaling factors that act to bridge any mechanistic gaps between the in vitro and in vivo milieu, and are typically based on determining the relative expression (or activity) or the difference in functional protein abundance between the in vivo and in vitro systems (Proctor et al., 2004). In particular, an expanding body of evidence has meant that judgments on the clinical progress of NCEs for certain P450-mediated drug-drug interactions can be reached by harnessing PBPK modeling strategies that predict its pharmacokinetic/drug-drug interaction liabilities (Jones et al., 2015; Wagner et al., 2015; de Zwart et al., 2016; Shebley et al., 2018). Furthermore, regulatory authorities have been developing guidance to ensure rigorous quality assurance is applied to NCE submissions that harness mechanistic PBPK modeling (https://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM531207.pdf; http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/2016/07/WC500211315.pdf).
Since protein standards historically have not been routinely available for membrane transporter expression absolute quantification, the capacity to scale transporter activity by IVIVE in order to predict the impact of transporter-mediated drug clearance within specific organs of a PBPK model has relied on transporter expression data from relative quantification approaches, i.e., mRNA expression quantification via reverse transcription polymerase chain reaction (PCR) or protein expression from immunoblotting to derive and apply relevant scaling strategies (Harwood et al., 2013; Neuhoff et al., 2013), while other related transporter IVIVE-PBPK models have required additional empirical scalars to ensure the model captures the clinical observations (Jones et al., 2012; Varma et al., 2012; Jamei et al., 2014). Recently, we undertook and reported on an extensive literature meta-analysis to establish 19 transporter protein abundances in the healthy Caucasian liver (Burt et al., 2016). Accompanying the meta-analysis was the development of an IVIVE strategy to harness interindividual variability in hepatic absolute transporter abundances (in picomoles) determined via the meta-analysis by using a unitless intersystem extrapolation factor for transporters (ISEF-T) within IVIVE. The drive to develop this strategy was the following: 1) a desire within the industry to develop more mechanistic scaling factors to facilitate model development for transporter IVIVE (Jones et al., 2015; Pan et al., 2016; Guo et al., 2018), and 2) the literary reporting of absolute transporter protein abundances in human livers utilizing burgeoning proteomics techniques (Heikkinen et al., 2015).
The capacity to scale transporter activity data obtained in relevant in vitro cell monolayers within PBPK models that describe region-specific intestinal transporter expression levels based on relative expression approaches has been demonstrated for intestinal efflux transporters such as P-glycoprotein (P-gp) (Neuhoff et al., 2013; Yamazaki et al., 2018). However, by building on the incorporation of absolute transporter abundance scaling of hepatic transporter activity (i.e., the ISEF-T approach), our aim was to perform an extensive meta-analysis of the expanding human intestinal transporter absolute abundance quantification literature in order to facilitate the development of an ISEF-T approach to scale transporter activity data in and along the gut. Furthermore, an appraisal of the literature utilizing relative quantification approaches was undertaken to determine region-specific expression of gut transporters, which could then be assimilated with that of absolute quantification–based studies. A similar rigor was applied to the gut transporter abundance meta-analysis as was done for that of the liver in terms of study exclusion criteria (Burt et al., 2016). We provide a meta-analysis of quantitative intestinal transport abundance data to employ the ISEF-T approach in IVIVE-PBPK.
Methods
Priming the Database: Identifying and Prioritizing Intestinal Transporters for Analysis.
There are more than 400 transporter genes identified in the human genome (César-Razquin et al., 2015). Therefore, prior to undertaking a systematic meta-analysis to establish region-specific transporter expression in the human intestine, it was important to primarily identify and prioritize transporter isoforms that show demonstrable expression in the human small and large intestines and possess the capacity to interact with drugs to potentially influence drug disposition. Based on analysis of the literature (via searching the PubMed electronic database; https://www.ncbi.nlm.nih.gov/pubmed), a database comprising 52 transporters relevant to the human intestine was collated, in which evidence was gathered and recorded on the transporter isoform(s) human intestinal expression and the methodology employed to quantify expression: membrane localization (apical, basolateral, or both), transporter function (i.e., uptake, efflux, or both), in vivo (human clinical) and in vitro evidence (cell monolayer studies) of interaction with drugs (substrate/inhibitor moieties), regulatory requirement/interest (i.e., the US Food and Drug Administration and European Medicines Agency), and focused groups such as the International Transporter Consortium. Given the available evidence, the transporters were ranked based on criteria such as robust evidence of mRNA transcription, protein expression, or known to be involved in intestinal drug absorption/disposition. Ultimately, the 16 highest ranked transporters were identified and are summarized in Table 1, including 10 transporters from the solute carrier family and six transporters from the ATP-dependent binding cassette superfamily, which were then prioritized for subsequent electronic literature searches and integration into an abundance database for meta-analysis.
The 16 transporters selected for inclusion into the meta-analysis after exclusion criteria were applied to determine region-specific transporter expression in the ADAM and M-ADAM models
Transporter Abundance Database.
A single overarching complete database was collated to contain published abundance data for the 16 prioritized transporters quantified in human intestinal tissue. The complete database included separate data sets, where the quantification of transporter abundances was performed using either absolute or relative approaches. Absolute transporter abundance data were typically quantified against a standard curve of a verified surrogate peptide(s) using quantitative targeted proteomics via liquid chromatography–tandem mass spectrometry or a quantitative western blot approach. In these assays, the transporter protein abundance is expressed in moles per mass of protein. For the relative transporter abundance data, quantification was typically performed using PCR or immunoblotting technology, where the abundance of the transporter was expressed relative to a housekeeper gene or protein. Original research articles were retrieved via searching the PubMed electronic database using combinations of the following keywords: human, intestinal, transporter, absolute, relative, protein, expression, abundance, and proteomics. The database including all available measurements was established (final literature search June 2017), with background information on the methods as well as donor demographics collated where provided. In cases where individual data were not directly reported, data were extracted via GetData Graph Digitizer (version 2.22, http://getdata-graph-digitizer.com) or authors were contacted directly to request individual donor data. A refined subdatabase was created through the use of various exclusion criteria. First, study methodologies were reviewed to ensure that absolute abundances were quantified using liquid chromatography–tandem mass spectrometry or quantitative western blot in total membrane (TM) fractions. For relative expression studies the same stringency for quantification in a TM fraction was not appropriate since numerous studies were included that required mRNA extraction and subsequent reverse transcription to cDNA for PCR-based expression quantification. Next, data in which it was stated that the human intestinal tissue was not from adult (aged <18 years), healthy (or macroscopically normal after histologic assessment), or Caucasian individuals were excluded. Any study in which pooling of sample matrices took place for mRNA, cDNA (Herrera-Ruiz et al., 2001; Seward et al., 2003), or microsomal samples for protein absolute abundance analysis (Nakamura et al., 2016) was not included since interindividual variability is lost when pooling as only mean with experimental error/deviation is, therefore, available. However, such data sets can be used to compare the results of meta-analyzed expression data of relevant transporters. For the relative expression studies, any study that did not normalize the target (i.e., transporter) gene or protein against a reference (or housekeeper gene/protein) within the same assay was excluded (Landowski et al., 2003; Kim et al., 2007). Exclusion occurred in instances where there was relative quantification of a transporter in a single region of the intestine (Hilgendorf et al., 2007) in which quantification only took place in the jejunum, hence normalization to other segments could not take place (see the procedural aspects given in Data Analysis: Determining Region-Specific Intestinal Transporter Expression via Meta-Analysis of Meta-Analysis). Finally, the sources of data were identified to ensure that duplicate measurements from the same tissue sample were not included in the refined database. Meta-analysis was then used to characterize the region-specific abundance of intestinal transporters in the refined database.
Data Analysis: Determining Region-Specific Intestinal Transporter Expression via Meta-Analysis.
Within the Simcyp Simulator the advanced dissolution absorption and metabolism (ADAM) and multilayer ADAM (M-ADAM) models, which constitute seven small intestinal segments (one duodenum, two jejunum, and four ileum segments) and a single segment representing the colon, contain the transporter expression specific to each intestinal segment (Jamei et al., 2009). The region-specific transporter expression is normalized relative to the proximal jejunum segment (jejunum I), as was previously described for ABCB1 (P-gp), ABCC2 [multidrug resistance-associated protein (MRP) 2], and ABCG2 (breast cancer resistance protein) using relative quantification approaches (Harwood et al., 2013). Hence, the meta-analysis was structured to take into account quantification of transporter abundance in each segment of the ADAM and M-ADAM models. Where there was insufficient information described in the study—for example, where samples were described as from the ileum and not described with greater precision to a specific region of the ileum—the expression data for that study were assigned with those expression values to each of the four ileum segments constituting ADAM and M-ADAM.
The meta-analysis comprised three data sets: 1) region-specific abundances determined from absolute abundance data, 2) region-specific abundances determined from relative expression data, and 3) region-specific abundances determined from a combination of absolute and relative data. For the meta-analysis, the region-specific abundances were determined after normalization to the weighted mean abundances in the reference segment of the proximal jejunum (i.e., jejunum I) (Harwood et al., 2013). For the relative abundance measurements that did not contain a jejunum sample, the values were initially scaled to colon or ileum and the relative average value was later used to combine all data relative to jejunum I.
In line with the previous meta-analysis (Harwood et al., 2013), where studies differentiated between colonic regions the transporter expression data were incorporated into the final analysis from those samples originating from the ascending colon, and where no suitable absolute abundance quantification data were available for a transporter, a jejunum I absolute abundance value of zero (in picomoles per milligram TM protein) was assigned. The meta-analysis proceeded to establish the region-specific abundance levels of that transporter based on relative transporter quantification methodology, where quantification was undertaken in the jejunum; therefore, normalization to jejunum I could be performed. In instances where abundance data generated via absolute or relative quantification methods were available for a transporter, the databases were combined within the meta-analysis framework.
After applying the exclusion criteria to the complete database, the collated abundance values for the healthy, Caucasian adult sub-database were combined for a given transporter to generate the weighted mean, geometric mean, S.D. and CV for the jejunum I segment of the extensive transporter (representing the wild type) phenotype based on the equations described previously for metabolizing enzymes (Perrett et al., 2007). The abundance values were further tested for between-study heterogeneity using the Cochran X2-based Q test (Perrett et al., 2007), whereby heterogeneity was apparent when a probability of P < 0.05 was returned.
The assigned CV was preferentially based on absolute abundance data when these were available for a transporter isoform, while for the majority of the other transporters analyzed, a CV based on relative expression data was required since absolute data were either not available or were excluded based on the aforementioned criteria. For SLC51A/B [organic solute transporter (OST)-α/β] and SLC10A2 [ileal bile acid transporter (IBAT)], additional considerations for CV determination were required. The rationale for their derivation is described in Results: Absolute Abundance Data Analysis, where unless otherwise stated the mean is provided in the text as mean ± S.D. for the abundance values (in picomoles per milligram of TM protein).
Total Membrane Protein Yield in the Small intestine and Colon.
To facilitate the scaling of in vitro activity data per picomole of transporter to the entire small intestine and colon requires that the human intestinal abundances, as determined in the aforementioned meta-analysis, are converted to picomoles per intestinal segment, thus enabling the calculation of segmental transporter activity (clearance). Since the meta-analysis of human intestinal abundances incorporated values reported as picomoles per milligram of TM protein, we sought to determine the small intestinal and colonic total membrane protein yield in milligrams.
Literature sources that specifically reported total membrane protein per intestine (TMePPI), which is related to small intestinal yield, and total membrane protein per colon (TMePPC) were sought. For each study, the TM protein yield for the specific intestinal segment from which the TM fractions were obtained was scaled to the duodenum, jejunum, and ileum dependent on the procedure, i.e., mucosal scraping (Tucker et al., 2012), mucosal crushing (Drozdzik et al., 2014), or enterocyte elution (Harwood, 2015). In instances where procedural losses during preparation of TM protein from tissue homogenates were available, these were taken into accounted in the final segmental yield calculations. The weighted mean values of the TMePPI and TMePPC are given in milligrams. The specific methodologies used to obtain segmental TM protein yield from each study are provided in the Supplemental Material in the section on the total membrane protein yield.
Results
Abundance Database.
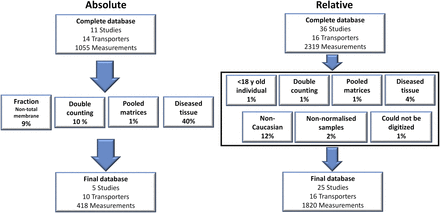
In this study, a total of 47 articles were recorded in the complete database of which 30 were accepted into the meta-analysis after exclusion criteria were applied. The complete database consisted of 3374 absolute and relative quantification measurements of transporter expression (see Supplemental Tables 2A and 2B for studies and sample quantification information). Of this complete database, 2238 relative- and absolute-based quantification measurements across all intestinal regions for 16 transporters matched our inclusion criteria and were thus included in the final data set for adult healthy Caucasians (Fig. 1). The final absolute database consisted of five independent studies (Tucker et al., 2012; Gröer et al., 2013; Oswald et al., 2013; Drozdzik et al., 2014; Harwood et al., 2015), and data from a Ph.D. program published in a thesis (Harwood, 2015), with the data linked to that published in Harwood et al. (2015). The most common reason for the exclusion of absolute abundance data was the use of samples from individuals with underlying disease conditions (40%), whereas for the relative abundance it was data from non-Caucasian samples (12%). Other reasons for exclusion of absolute abundance data were the reporting of data from duplicate samples (Brück et al., 2017) and quantification in samples other than TM fractions (9% of complete absolute database) (Wiśniewski et al., 2015; Vaessen et al., 2017), while the other criteria for exclusion in both the absolute and relative data sets constituted a relatively minor component (Fig. 1; Supplemental Table 1). There was limited information available on an individual’s phenotype status; therefore, no studies were excluded for possessing nonextensive-transporter phenotype samples. The samples quantifying absolute abundances and excluded due to underlying disease were due to individuals who were morbidly obese with a body mass index >30 (Miyauchi et al., 2016) and those samples classified as possessing adenoma or cancer (Wiśniewski et al., 2015), while for the relative abundance quantifications, Crohns and ulcerative colitis samples (Thibault et al., 2007) required that these were excluded from the refined database. Non-Caucasian ethnicity samples were excluded since they incorporated abundance data from Japanese individuals (Hinoshita et al., 2000; Terada et al., 2005).
Exclusion criteria applied to the complete database for the absolute (left) and relative (right) abundance data. Percentages for each exclusion criterion refer to the fraction of the samples in the complete database that were excluded on its basis.
Of the 16 transporters on which the final meta-analysis was based (Table 1), suitable absolute data could not be obtained for six transporters at the time of this meta-analysis due to one or more of the criteria outlined in Fig. 1. These transporters were SLC16A1 (monocarboxylate transporter 1), SLC22A4 [organic cation transporter (OCT), type 1 (OCTN1)], SLC2A2 [glucose transporter 2 (GLUT2)], ABCC1 and ABCC4 (MRP1 and MRP4, respectively), and SLCO4C1 [organic anion transporting polypeptide (OATP) 4C1]. Where absolute abundance data were not available for any transporter, the relative quantification data were used to determine the region-specific abundance in the intestine.
Absolute Abundance Data Analysis.
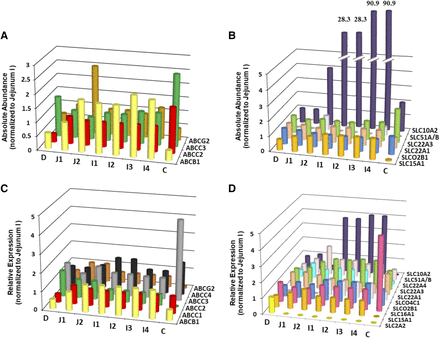
A summary of the absolute abundances for the 10 transporters in the healthy Caucasian adult proximal jejunum, assigned as jejunum I in the ADAM model, is provided in Table 2. The solute carrier apical uptake carrier peptide transporter 1 (PepT1) showed the highest abundance in the human jejunum I samples with a weighted mean abundance of 3.69 ± 1.5 pmol/mg TM protein (n = 11). The highest abundance the ATP-binding cassette transporter was MRP2 with 0.86 ± 0.58 pmol/mg TM protein (n = 11). In this analysis, the mean jejunum I abundance for OST-α/β was taken from analysis of distal rather than proximal jejunum due to the lack of quantification in proximal jejunum regions. OST-α/β is a dimeric protein conferring functionality when both α and β subunits coalesce (Seward et al., 2003). However, absolute quantification methods typically endeavor to quantify the abundance of each subunit separately (Harwood, 2015). For this analysis, the α subunit was used for OST-α/β abundance in jejunum I since the α subunit is considered to be the limiting component in conferring activity for this dimer (Sun et al., 2007). With the translation of in vitro transporter activity through the ADAM and M-ADAM models, the variability (CV) for a given transporter was assigned for jejunum I only, and this variability was applied and propagated through all of the segments of the gastrointestinal tract (Neuhoff et al., 2013). The highest variability was also associated with OST-α/β, having CV values of 99% (Table 2), which was derived from the combination of the relative and absolute data analysis for the jejunum samples due to the limited number of samples from the absolute analysis (i.e., n = 1 in jejunum II). The levels for IBAT are negligible in the proximal small intestine (Gröer et al., 2013; Drozdzik et al., 2014), which may give rise to exaggerated interindividual variability due to analytical imprecision at such low levels of abundance. Hence, for this analysis the CV values were assigned from PCR-based jejunum mRNA analysis (Hilgendorf et al., 2007). In the final database, heterogeneity in absolute abundance values was found for MRP2 in the duodenum (P = 0.049) and ileum II (P = 0.047) segments. There was no between-study heterogeneity found for the other transporters within the absolute abundance database. The region-specific abundance based on the absolute abundance data set, once normalized to jejunum I, is provided in Fig. 2, A and B, with values given in Supplemental Table 3. Figure 3 shows the relative proportion of the abundance for each transporter in the final database after performing a simulation in 2000 North European Caucasians (the values are provided in Supplemental Table 4).
The weighted mean, coefficient of variation, and geometric mean of total membrane protein abundance of drug transporters in the proximal jejunum (jejunum I) obtained from the meta-analysis of measurements in tissue of healthy Caucasian adults
Absolute abundance quantification of ATP-binding cassette (ABC) transporters normalized to the jejunum I segment (A) and solute carrier (SLC) family transporters (B) in all intestinal segments representing the ADAM model (C, colon; D, duodenum; I1–I4, ileum I1–I4 segments; J1 and J2, jejunum I and II segments). Relative abundance quantification of ABC transporters normalized to the jejunum I (C) and SLC transporters (D) in all intestinal segments representing the ADAM and M-ADAM models. The bars represent the weighted mean normalized abundance of each transporter. Representative values depicted in this figure are provided in Supplemental Table 3. Where no abundance data were available a zero value was assigned. This is represented in histogram B for SLC51A/B in all segments except jejunum II and ileum IV and in histogram D for SLC2A2 in all segments except duodenum, SLCO4C1 duodenum, and jejunum I and II. The values provided above the bars for SLC10A2 in histogram B represent the scaled up expression relative to the break point [(//), i.e., 5] executed for these values.
Intestinal drug transporter pies. Proximal jejunum (jejunum I) ATP-binding cassette (ABC) (A) and solute carrier (SLC) and OATP (SLCO) (B). Distal ileum (ileum IV) ABC (C) and SLC and OATP (SLCO) (D). Protein abundance in the final subdatabase for each transporter family transporters as a percentage of the total abundance (in picomoles) of the region shown after performing a simulation with 2000 North European Caucasians.
Relative Abundance and Combining Relative and Absolute Abundance Data Analysis.
Summaries of the studies recorded and the region-specific abundances based on analysis utilizing relative abundance quantification methodology are provided for 16 transporters in the healthy Caucasian adult in Fig. 2, C and D, Supplemental Table 2B, and Table 3. With the exception of IBAT (Fig. 2, A and D), there is good consistency when comparing the region-specific abundances determined from the absolute or relative quantification methodology (Supplemental Table 3). The differences observed for this protein between methodologies may be due to the ability of the highly sensitive targeted proteomics analysis used to derive low abundances for this protein in the proximal jejunum (∼0.01 pmol/mg total membrane protein) (Gröer et al., 2013; Drozdzik et al., 2014). Nevertheless, an increasing gradient of expression peaking in the terminal ileum is expected for this protein. For GLUT2, relative quantification data were only available in the duodenum; therefore, normalization to jejunum I was not possible and a relative expression value of 1 was assigned across regions. Although not shown specifically here, for the studies excluded due to ethnicity, i.e., Japanese samples (Hinoshita et al., 2000; Terada et al., 2005), the mRNA expression normalized to glyceraldehyde-3-phosphate dehydrogenase in the colon was ranked MRP3 > MRP1 = MDR1 > MRP2 (where MDR1 is defined as multidrug resistance 1), and in the final Caucasian analysis (Table 3) the ranking was similar once normalized to jejunum I with the exception of MRP1 and MDR1, showing a more pronounced difference in the Japanese data set (MRP3 > MRP1 > MDR1 > MRP2) (Hinoshita et al., 2000). While the Japanese data for the region-specific mRNA expression were fairly similar for MDR1, OCTN1, and OCT1, there was a more distally distinct decrease in expression compared to the Caucasian analysis for PepT1 (Table 3), in which there was a more uniform distribution along the small intestinal segments (Terada et al., 2005). In the Japanese samples, OCT3 generally showed lower regional mRNA expression than Caucasians; however, its region-specific expression showed similar trends to Caucasians (Table 3) (Terada et al., 2005).
Final region-specific abundances along the gastrointestinal tract normalized to jejunum I based on relative abundance quantification or by combining abundance data obtained from relative and absolute quantification methodology
To enhance the rigor of the meta-analysis and facilitate the incorporation of an increasing number of relevant and new transporters into the ADAM and M-ADAM models, the absolute and relative quantification results for transporters were combined to obtain region-specific abundances and interindividual variability specifically for seven transporters (Tables 1 and 3). Furthermore, the final relative abundances incorporated into the ADAM and M-ADAM models are provided in Table 3. Irrespective of whether the meta-analysis for a transporter used a combined approach, or only relative quantification methodologies were available, the transporters displaying significant region-specific expression compared with the proximal jejunum (Table 3) included the following: IBAT that showed a 100-fold or more greater expression in the terminal ileum, ABCC3 (MRP3) that showed >5-fold higher expression in the colon and >2-fold higher in the duodenum, and OCTN1 and SLC22A1 (OCT1) that showed >2-fold lower expression in the colon. For all other evaluated transporters, the region-specific expression was relatively uniform across the segments with no transporter displaying expression greater or less than 2-fold higher than in the proximal jejunum.
Intestinal and Colon Total Membrane Protein Yield.
The yields of TM protein in the intestine (TMePPI) and colon (TMePPC) were determined from three studies (Tucker et al., 2012; Drozdzik et al., 2014; Harwood, 2015). The yields based on the study by Drozdzik et al. (2014) were obtained via personal communication from Dr. Stefan Oswald (University of Greifswald, Greifswald, Germany). The data set consisted of 35 sample measurements, n = 14 duodenal (Tucker et al., 2012), n = 5 jejunum and n = 3 ileum (Harwood, 2015), and n = 7 colon (Drozdzik et al., 2014; Harwood, 2015), where the age and gender distribution (when known) was 24–72 years, with a minimum of two females. The small intestinal TM protein yield (n = 6) from Drozdzik et al. 2014 was provided as a lumped value covering the entire small intestinal region, as was the colon (n = 6) from the same study. The five jejunum samples measured from Harwood (2015) consisted of one sample from the proximal jejunum with the remainder from the distal jejunum (one female; 41–62 years). Since each study did not contain specific TM protein yield data for each segment, the capacity to determine TMePPI and TMePPC required several conversions and assumptions in order to estimate yields in regions that were not measured experimentally (Supplemental Material, see the section on the total membrane protein yield). The weighted mean (± S.D.) values from the three studies were 2737 ± 1807 mg for TMePPI and 112 ± 37 mg for TMePPC, which were used to calculate the absolute abundance of protein in picomoles per milligram TM protein from the meta-analysis to picomole concentrations.
Discussion
The heightened recognition that PBPK models play a role in evaluating the mechanisms responsible for drug pharmacokinetics at industrial and regulatory levels has driven the demand to quantify key drug and physiologic elements precisely and accurately in PBPK models. The relative expression factor (REF) approach provided a means to scale transporter activity from cell monolayers in various regions of the intestine (Neuhoff et al., 2013). Although practical, these models are not as sophisticated as those for P450s, in which the intersystem extrapolation factor scalar corrects for activity differences per unit of enzymes in the liver versus recombinant systems (Proctor et al., 2004). The increasing utilization of methodologies to quantitatively determine a protein’s absolute abundance within a biologic system has led to efforts from developers of PBPK model platforms to harness these data within a physiologic framework. The ISEF-T approach permits transporter-specific scaling of in vitro kinetics based on a protein’s molar concentration within an individual’s organ (Burt et al., 2016). Given the increasing availability of region-specific intestinal transporter expression data from the relative and absolute quantification approaches, a rigorous meta-analysis is provided to obtain region-specific transporter abundances and variability in healthy adult Caucasians.
Literary evidence was evaluated to prioritize intestinal transporters involved in drug disposition for inclusion into the meta-analysis. To accurately construct PBPK models that can assess the impact of transporter proteins, substantiating their intestinal expression, function, and localization on the enterocyte plasma membrane are imperative. In vitro studies were required to assess the plasma membrane localization and functional aspects for certain transporters, while intestinal transporter expression was confirmed primarily via immunoblotting, quantitative proteomics, immunohistochemistry, and mRNA expression. The clinical pharmacokinetic relevance was also considered (http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/2012/07/WC500129606.pdf; https://www.fda.gov/downloads/drugs/guidances/ucm292362.pdf) (Zamek-Gliszczynski et al., 2018). For the majority of transporters (Table 1), intestinal protein expression was unequivocal (P-gp, MRP2, MRP3, breast cancer resistance protein (BCRP), OATP2B1, IBAT, PepT1, monocarboxylate transporter 1, OCT1, and OCT3). For MRP1, OCTN1, MRP4, and OST-α/β, where proteomics data were limited to a single study or intestinal region (Harwood, 2015; Wiśniewski et al., 2015; Nakamura et al., 2016), supporting evidence related to other protein quantification methods or mRNA expression was sought prior to ranking for inclusion. For OATP4C1, its capacity to transport the P-gp probe digoxin across the basolateral membrane of the renal proximal tubule cell is implicated (Mikkaichi et al., 2004), while transcriptional information supports potential expression in the small intestine and immunohistochemistry demonstrates basolateral membrane expression in colon enterocytes (Hilgendorf et al., 2007; Bourgine et al., 2012; Kleberg et al., 2012). OATP1A2 (one of the initial 52 transporters evaluated) is of potential pharmacokinetic relevance; however, several studies show intestinal mRNA and protein levels are absent or negligible (Supplemental Table 1). Hence, this transporter is not included in the final meta-analysis. GLUT2 is considered in the model as a potential drug target for obesity and diabetes since jejunal GLUT2 is highly abundant in morbidly obese individuals (Miyauchi et al., 2016). In obesity, GLUT2 translocates from the enterocyte’s basolateral membrane in healthy individuals to the apical membrane (Ait-Omar et al., 2011). Understanding a transporter’s location and function is crucial to constructing the appropriate model structure and algorithms to accurately scale transporter activity. There is conflicting information regarding the enterocyte localization of OCT1, in which earlier studies implicated the lateral/basolateral membrane localization (Müller et al., 2005; Giacomini et al., 2010) but a later study that combined immunocytochemistry and functional transporter assays using the OCT1-specific substrate pentamidine concluded that apical uptake predominated (Han et al., 2013). Hence, OCT1 is assigned as an apical membrane uptake transporter (Table 1). The enterocyte localization of OATP2B1 has also been under scrutiny recently (Keiser et al., 2017). Early studies implicated apical localization on human intestinal sections and Caco-2 cells (Kobayashi et al., 2003; Sai et al., 2006). However, recent proteomic investigations demonstrated markedly higher OATP2B1 expression in basolateral compared with apical membrane fractions; however, accompanying immunostaining revealed less emphasis on basolateral localization (Keiser et al., 2017). Given these findings, OATP2B1 expression in both apical and basolateral membranes is plausible, thus OATP2B1 is assigned as an apical uptake transporter (Table 1). However, the switching of transporter localization and function in the model is possible, thus OATP2B1 could represent a basolateral uptake transporter.
To ascertain region-specific absolute transporter abundances via meta-analysis for the prioritized 16 transporters, a complete database was compiled cataloging 3374 measurements across all intestinal regions. To define the healthy Caucasian adult intestinal transporter expression a refined database was curated, where several exclusion criteria were applied such that the largest proportion of measurements excluded were that of non-healthy samples (Fig. 1). Disease can affect transporter expression (Evers et al., 2018); therefore, distinguishing between those samples that are directly affected by disease is critical. However, the challenge with curating such a data set is that a routine means of obtaining intestinal samples to quantify protein expression is from individual’s undergoing surgical intervention for an array of complications. In such cases, if studies classified their samples as macroscopically normal it was assumed that the tissue’s protein expression was unaffected by the disease, thus these measurements were incorporated into the refined database.
The ADAM and M-ADAM models scale intestinal transporter activity in a region-specific manner after normalization against the reference jejunum I segment in both the ISEF-T and REF approaches (Neuhoff et al., 2013). Therefore, the primary goal is to determine the jejunum I absolute transporter protein abundance and its associated interindividual variability (Table 2). For certain transporters, it was necessary to obtain the CV values from absolute and relative data sets, since there were insufficient jejunum-based absolute abundance quantifications (OST-α/β) or jejunum-mRNA quantification was being considered (IBAT). For all other intestinal segments where data were available, the weighted mean transporter abundances were determined and normalization to the jejunum I abundance (picomoles per milligram TM protein) was performed. Between-study heterogeneity was not as evident as in the healthy Caucasian adult liver absolute abundance meta-analysis (Burt et al., 2016), with MRP2 being the only transporter displaying this tendency in two segments. This may be due to less biologic variability between the studies or lower sample numbers available to distinguish heterogeneity than for the liver. Due to the limited availability of measurements that can be directly attributed to a given donor, it was not possible to perform any correlation analysis with respect to age and gender.
Although performing region-specific meta-analysis using studies quantifying transporter protein or mRNA expression using relative quantification techniques cannot directly inform us of the absolute levels of transporter abundances within a sample, it does provide a relatively large number of measurements, which provide robust region-specific expression information for each transporter after normalization to jejunum I when combined with the absolute data set. Except for IBAT, the relative or absolute quantification techniques showed limited region-specific transporter differences. This provided us with the confidence to use both absolute and relative data sets in order to obtain the final region-specific abundances for incorporation into a healthy Caucasian population. Alone, the relative expression data set was instrumental in providing region-specific expression data for six transporters (Tables 1 and 3), allowing the REF approach to be employed even if absolute abundances were not available.
Absolute abundance data were available from whole tissue homogenate, TM fractions, and plasma membrane fractions. However, only data obtained from TM fractions were included in the final database, since corresponding protein yield values enabling the conversion of abundance values to picomoles per transporter/intestinal segment are reported here for the first time. Derivations of the intestinal (TMePPI = 2737 ± 1807 mg) and colonic (TMePPC = 112 ± 37 mg) total membrane protein yields were based on three studies in which the primary focus of each study was not the determination of these parameters (Tucker et al., 2012; Drozdzik et al., 2014; Harwood, 2015). Therefore, the application of several assumptions based on determination of intestinal/mucosal tissue yields (Paine et al., 1997) and intestinal mucosal cylindrical surface areas facilitated the determination of the yields being dependent on whether the mucosa or enterocytes specifically were processed (Supplemental Material, see the section on the total membrane protein yield). The TMePPI is in reasonable agreement with the small intestine microsomal protein yield of 2978 mg, which is anticipated given the methodological similarity in obtaining microsomes (Paine et al., 1997) or TM fractions (Tucker et al., 2012) using differential centrifugation. It is difficult to gauge the physiologic plausibility of the TMePPC since there are no equivalent colonic microsomal data published for comparative purposes; however, a lower TMePPC is expected than TMePPI, given the highly folded structure of the small intestinal mucosa compared with the colon. Further dedicated studies are sought to investigate intestinal membrane protein yield.
This is the first in-depth systematic analysis of intestinal region-specific transporter expression based on absolute abundance quantification methods. The expression data derived herein provide additional flexibility in modeling region-specific active transport processes for 16 transporters expressed in the enterocyte utilizing both relative (REF) and absolute abundance (ISEF-T) scaling approaches. To use the ISEF-T approach, increasing focus on measuring in vitro transporter kinetics combined with absolute transporter abundances (Meng et al., 2017a,b) is required. Routine updates to the database are required when applicable data become available (Drozdzik et al., 2018), and further studies are warranted to enhance our ability to translate absolute abundance quantification data into mechanistic models, while similar analyses are required for other ethnicities and disease models.
Acknowledgments
We thank Dr. Stefan Oswald (University of Greifswald, Greifswald, Germany) for personally communicating the total membrane protein yield data from the Drozdzik et al., (2014) study, Dr. Iain Gardner for productive discussions, and Dr. Gopal Pawar (formerly of Certara UK Ltd., Simcyp Division) for assisting with the initial selection of transporters for inclusion in the database. We also thank Eleanor Savill for assistance in formatting the manuscript for submission.
Authorship Contributions
Participated in research design: Harwood, Zhang, Pathak, Neuhoff.
Conducted experiments: Harwood, Zhang, Pathak, Neuhoff.
Performed data analysis: Harwood, Zhang, Pathak, Neuhoff.
Wrote or contributed to the writing of the manuscript: Harwood, Zhang, Pathak, Neuhoff.
Footnotes
- Received March 15, 2019.
- Accepted April 30, 2019.
↵1 Current affiliation: Quotient Sciences, Nottingham, United Kingdom.
This research was funded by Certara UK Ltd. (Simcyp Division).
This work was presented as a conference proceeding [Abstract and Poster (208)] at the 21st North American ISSX Meeting, September 2017, Providence, RI. The title of the Abstract was “Abundance & relative segmental expression of intestinal transporters in caucasians: a meta-analysis.” This can be viewed at the following website: http://issx.confex.com/issx/21NA/webprogram/Paper37982.html and in Drug Metabolism and Pharmacokinetics 33(1):S88–S89 DOI: 10.1016/j.dmpk.2017.11.289.
↵
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
Abbreviations
- ADAM
- advanced dissolution absorption and metabolism
- GLUT2
- glucose transporter 2
- IBAT
- ileal bile acid transporter
- ISEF-T
- intersystem extrapolation factor for transporters
- IVIVE
- in vitro-to-in vivo extrapolation
- M-ADAM
- multilayer advanced dissolution absorption and metabolism
- MDR1
- multidrug resistance protein 1
- MRP
- multidrug resistance-associated protein
- NCE
- new chemical entity
- OATP
- organic anion transporting polypeptide
- OCT
- organic cation transporter
- OST
- organic solute transporter
- P450
- cytochrome P450
- PBPK
- physiologically based pharmacokinetic
- PCR
- polymerase chain reaction
- PepT1
- peptide transporter 1
- P-gp
- P-glycoprotein
- REF
- relative expression factor
- TM
- total membrane
- TMePPC
- total membrane protein per colon
- TMePPI
- total membrane protein per intestine
- Copyright © 2019 The Author(s).
This is an open access article distributed under the CC BY Attribution 4.0 International license.