Abstract
Carboxylesterase (CES) 1 is the most abundant drug-metabolizing enzyme in human livers, comprising approximately 1% of the entire liver proteome. CES1 is responsible for 80%–95% of total hydrolytic activity in the liver and plays a crucial role in the metabolism of a wide range of drugs (especially ester-prodrugs), pesticides, environmental pollutants, and endogenous compounds. Expression and activity of CES1 vary markedly among individuals, which is a major contributing factor to interindividual variability in the pharmacokinetics (PK) and pharmacodynamics (PD) of drugs metabolized by CES1. Both genetic and nongenetic factors contribute to CES1 variability. Here, we discuss genetic polymorphisms, including single-nucleotide polymorphisms (SNPs), and copy number variants and nongenetic contributors, such as developmental status, genders, and drug-drug interactions, that could influence CES1 functionality and the PK and PD of CES1 substrates. Currently, the loss-of-function SNP G143E (rs71647871) is the only clinically significant CES1 variant identified to date, and alcohol is the only potent CES1 inhibitor that could alter the therapeutic outcomes of CES1 substrate medications. However, G143E and alcohol can only explain a small portion of the interindividual variability in the CES1 function. A better understanding of the regulation of CES1 expression and activity and identification of biomarkers for CES1 function in vivo could lead to the development of a precision pharmacotherapy strategy to improve the efficacy and safety of many CES1 substrate drugs.
SIGNIFICANCE STATEMENT The clinical relevance of CES1 has been well demonstrated in various clinical trials. Genetic and nongenetic regulators can affect CES1 expression and activity, resulting in the alteration of the metabolism and clinical outcome of CES1 substrate drugs, such as methylphenidate and clopidogrel. Predicting the hepatic CES1 function can provide clinical guidance to optimize pharmacotherapy of numerous medications metabolized by CES1.
Introduction
Carboxylesterase (CES) 1 is a phase I drug-metabolizing enzyme (DME) responsible for 80%–95% of total hydrolytic activity in the liver (Imai et al., 2006); it metabolizes a wide range of drugs, pesticides, environmental pollutants, and endogenous compounds, including lipid esters (Table 1). CES1-mediated metabolism can lead to the biotransformation of a pharmacologically active drug into its inactive metabolite, as exemplified by methylphenidate hydrolysis in the liver. CES1 also plays an important role in activating prodrugs since most ester-containing prodrugs are exclusively dependent on CES1 for their activation. The clinical relevance of CES1 has been well demonstrated in various clinical trials with oseltamivir, methylphenidate, and clopidogrel (Zhu et al., 2008; Tarkiainen et al., 2012; Lewis et al., 2013; Jiang et al., 2016). Recent studies have also revealed that CES1 acts as a cholesteryl ester hydrolase in lipid metabolism in human macrophages and hepatocytes and suggest CES1 as a potential drug target for the treatment of metabolic diseases, such as diabetes and atherosclerosis (Dolinsky et al., 2004; Zhao et al., 2007; Ghosh et al., 2010; Ross et al., 2010; Lian et al., 2018b).
List of CES1 substrates
Importance of CES1 in Drug Metabolism
CES1 plays an important role in metabolizing many clinically significant medications, especially the ester-prodrugs (Table 1). A prodrug refers to an inactive drug molecule that needs to be enzymatically biotransformed in vivo to its active metabolite to produce its intended pharmacological effect (Rautio et al., 2008). Prodrug design offers an attractive method to overcome the issue of low bioavailability for Biopharmaceutics Classifications System (BCS) class III drug molecules. Drug molecules can be categorized into four BCS classes based on permeability and solubility, and a BCS class III substance is a hydrophilic compound with low permeability and high solubility (Shah and Amidon, 2014). In particular, hydrophilic compounds with –OH or –COOH functional groups usually have difficulty being absorbed into the body, and drug developers often mask these functional groups using an ester-prodrug design. The prodrug market has been growing: 20% of drugs approved in 2015 were prodrugs compared with ∼6% of all currently approved drugs (Rautio et al., 2017).
Two major assumptions behind the ester-prodrug design are that prodrugs are rapidly activated via unspecific esterases in the body and that the interindividual variability in activating a prodrug is clinically insignificant. These incorrect assumptions may have stemmed from the fact that many hydrolytic enzymes exist in the body, such as CES1, CES2, acetylcholinesterase, butyrylcholinesterase, paraoxonases, and arylesterase. However, these hydrolases differ in their tissue-specific expression, cellular localization, and, most importantly, substrate selectivity (Fukami and Yokoi, 2012). In humans, CES1 is highly abundant in the liver and expressed to a lesser extent in the lung and brain; CES1 expression is considered negligible in the human intestine, kidney, and plasma. CES1 is substrate-selective toward carboxyl esters with a large ethyl group and a small alcohol group. In comparison, CES2, another major carboxylesterase in humans, is highly expressed in the intestine, kidney, and liver and is more efficient at metabolizing compounds with a small ethyl group and a large alcohol group (Jewell et al., 2007). Numerous in vivo and in vitro studies have demonstrated the specificity of CES1, and many CES1 substrates cannot be metabolized by other esterases (Table 3).
CES1 expression and activity vary significantly among individuals (Wang et al., 2016b); this variability could result in treatment failure and unexpected adverse effects of CES1 substrate drugs. A better understanding of the genetic and nongenetic factors contributing to CES1 variability will improve the design and clinical use of many drugs that are metabolized (deactivated/activated) by CES1.
Pharmacogenetics of Drug-Metabolizing Enzymes
Traditionally, fixed-dose regimens have been used for most medications. However, different individuals taking the same dose of medication do not necessarily achieve the same drug exposure and, hence, drug response. More individualized, patient-centered dosing regimens have been developed based on a patient’s characteristics, such as renal clearance, liver function, body weight, and surface area (DiPiro, 2017). In addition, genetic polymorphisms of DMEs have been found to play an important role in the response to pharmacotherapy, and pharmacogenomics has been increasingly used in the clinic to improve the efficacy and safety of drug treatment. DMEs serve to primarily detoxify digested xenobiotics through four general mechanisms: hydrolysis (e.g., carboxylesterase), reduction (e.g., carbonyl reductase), oxidation (e.g., cytochrome P450), and conjugation (e.g., UDP-glucuronosyltransferase) (Foti and Dalvie, 2016). The expression and activity of DMEs vary significantly among individuals, and studying pharmacogenomics of DMEs is one means of better understanding interindividual variability in the pharmacokinetics (PK) and pharmacodynamics (PD) of a drug. For example, the active metabolite of irinotecan, SN-38, is primarily metabolized by the enzyme UDP-glucuronosyltransferase family 1 member A1 [(UGT1A1) enzyme] (Ando et al., 2000). If a patient carries the common UGT1A1*28 polymorphism, the decrease it causes in UGT1A1 enzymatic activity would impede the metabolism of SN-38, leading to the accrual of toxic concentrations. Accordingly, the Food and Drug Administration (FDA) recommended that patients with UGT1A1*28/*28 start irinotecan at a lower dose (Innocenti et al., 2004). However, given that both genetic and environmental factors contribute to DME function, we should also pay close attention to nongenetic contributors when studying the variability of DMEs.
CES1 Pharmacogenetics
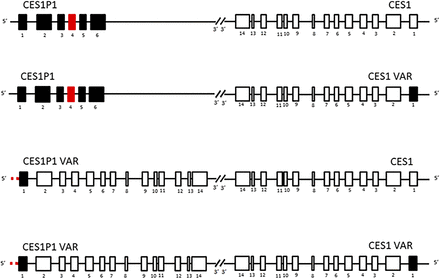
Although CES1 plays a critical role in the metabolism of many clinically important medications, CES1 pharmacogenetics is understudied relative to other major DMEs [e.g., cytochrome P450 (CYPs)]. CES1 is encoded by the CES1 gene and consists of 14 exons located on chromosome 16q13-q22.1. CES1 VAR is a variation of the CES1 gene that differs in exon 1 DNA sequences and has an average minor allele frequency (MAF) of 17%. Although one study claimed that CES1 VAR mRNA was undetectable (Fukami et al., 2008), an in vitro human liver study showed that the protein expressions of CES1 and CES1 VAR were not statistically different (Wang et al., 2016b). CES1P1 is a pseudogene due to a premature stop codon in exon 4 and lies tail-to-tail with CES1 (Fig. 1) (Wang et al., 2016b). Interestingly, a CES1P1 variant named CES1P1 VAR is a functional coding gene with a DNA sequence identical to CES1 VAR. However, the transcription efficiency of CES1P1 VAR is only 2% of that of CES1 because of the transcription factor specificity protein 1 and the enhancer-binding protein, CCAAT-enhancer-binding protein α, preferring to bind to the CES1 promoter over the CES1P1 VAR promoter (Hosokawa et al., 2008; Yoshimura et al., 2008). Because of the existence of the CES1 VAR and CES1P1 VAR variants, four CES1/CES1P1 haplotypes can be formed (Fig. 1). In addition to these structural variations, there are over 7000 CES1 single-nucleotide polymorphisms (SNPs) registered in the National Center for Biotechnology Information SNP database, and approximately 300 of them have MAFs over 1%. These common CES1 variants (MAF >1%) are distributed in various regions of the gene, including 13 in 5′– and 3′–untranslated regions, 14 in exons, and 308 in introns. Of the exonic SNPs, 12 are nonsynonymous SNPs, and two are synonymous SNPs. In the following section, we discuss the clinical findings and mechanistic bases of functional CES1 variants identified to date.
CES1 gene structure and haplotypes. CES1 gene consists of 14 exons located on chromosome 16q13-q22.1, and CES1P1 is a pseudogene, lying tail-to-tail with CES1. CES1, CES1P1, and their variants CES1 VAR and CES1P1 VAR form four major haplotypes. Red represents where stop codon is located. Transcription efficiency of CES1P1 VAR is approximately 2% of CES1.
Pharmacogenetics of the First Loss-of-Function CES1 Variant G143E (rs71647871)
In SNP notation, G143E indicates an amino acid change from glycine to glutamic acid at amino acid position 143. G143E is also termed 428G>A, indicating that the nucleotide guanine is changed to adenine at position 428 of the CES1 mRNA (DiPiro, 2017). The MAF of G143E is 3.7%, 4.3%, and 2%, in White, Hispanic, and African American populations, respectively, whereas the SNP is extremely rare in Asian populations (Zhu et al., 2008; Suzaki et al., 2013a).
G143E is a nonconservative amino acid substitution located near the active-site triad residues of CES1 (serine 221, glutamic acid 354, and histidine 468). Serine hydrolases share similar catalytic mechanism involving 1) nucleophilic attack from oxygen in the serine residue on a substrate ester bond, 2) formation of a tetrahedral intermediate wherein the deprotonated oxygen is stabilized via an oxyanion hole, 3) formation of an acyl-enzyme intermediate, and 4) water-catalyzed hydrolysis (Satoh and Hosokawa, 2006). For CES1 to maintain its enzymatic function, the catalytic triad and oxyanion hole need to be conserved (Zhu et al., 2008; Arena de Souza et al., 2015). The change from glycine (hydrophobic residue) to glutamic acid (electrostatic residue) at codon 143 disrupts the hydrophobicity needed for the oxyanion hole (Gly 141-131), resulting in a complete loss of function of CES1. The G143E is only CES1 SNP that has been subjected to in vitro kinetics studies in which the variant exhibited null catalytic activity on all tested CES1 substrates except for oseltamivir (Table 2). The Vmax of G143E on oseltamivir hydrolysis was 37 nmol/min per milligram with catalytic efficiency of 17.2 μl/min per milligram protein—this was approximately 16% of wild-type CES1 catalytic efficiency (Zhu and Markowitz, 2009).
In vitro kinetics of wild-type CES1 in human liver S9 fractions (HLS9)
Discovery of G143E and Its Impacts on Methylphenidate PK and PD.
G143E is the first loss-of-function (LOF) variant known for CES1 and was originally discovered in a methylphenidate (Ritalin) PK study in healthy volunteers. Methylphenidate is a central nervous system stimulant and the most commonly prescribed medication for attention deficit hyperactivity disorder (ADHD) treatment. Methylphenidate has high abuse potential when used with alcohol (COTEMPLA XR-ODT(TM), 2017). Its drug product comes as a racemic mixture of d- and l-methylphenidate hydrochloride; d-methylphenidate is approximately 10 times more pharmacologically potent than l-methylphenidate (Heal and Pierce, 2006).
Methylphenidate is metabolized by de-esterification via CES1 to ritalinic acid, an inactive metabolite that accounts for approximately 80% of the recovered dose in human urine (Fig. 2) (Laizure et al., 2013; COTEMPLA XR-ODT(TM), 2017). In 2007, a prospective single-dose (0.3 mg/kg) PK study was conducted in 20 healthy volunteers to examine the drug-drug interaction (DDI) between methylphenidate and alcohol (Patrick et al., 2007). During this study, the researchers unexpectedly found a participant that showed significantly elevated pharmacokinetic parameters [e.g., area under the curve (AUC), Cmax] of methylphenidate. Specifically, dl-methylphenidate Cmax was seven times higher and l-methylphenidate Cmax was 100-fold higher in this poor metabolizer compared with the rest of the participants. Later analysis found that this poor metabolizer carried the G143E polymorphism in CES1 and the D260fs polymorphism in CES1P1 (Zhu et al., 2008). This study also concluded that though CES1 metabolism is substantially stereoselective toward l-methylphenidate, d-methylphenidate metabolism is also significantly impacted by CES1 dysfunction.
D-methylphenidate comes as a single active ingredient (Focalin) or in combination with l-methylphenidate (racemic mixture) (Ritalin). D-methylphenidate is approximately 10 times more pharmacologically potent than l-methylphenidate, whereas l-methylphenidate is a better CES1 substrate. Ethylphenidate can be formed via transesterification with ethanol.
Following the discovery of the G143E variant, a retrospective study was conducted to examine the methylphenidate response in Hungarian patients with ADHD; G143E (n = 7) carriers and noncarriers (n = 115) were compared. Even though the CES1 genotype could not explain the entire interindividual variability between responders (n = 90) and nonresponders (n = 32), the study demonstrated an association between G143E polymorphism and methylphenidate dose reduction: five responders who had the G143E polymorphism required lower doses of methylphenidate for symptom reduction (0.410 vs. 0.572 mg/kg, P = 0.022) (Nemoda et al., 2009). In 2017, a healthy volunteer study confirmed the significance of G143E in the PK of methylphenidate. In this open-label, prospective clinical trial (n = 22), study participants carrying the G143E SNP (n = 6) had approximately 152.4% higher median AUC of d-methylphenidate (53.3 ng × ml−1 × h−1) than the noncarrier group (21.4 ng × ml−1 × h−1) (P < 0.0001) (Stage et al., 2017a).
The above studies suggest that G143E carriers may be at high risk of being exposed to a toxic methylphenidate concentration. This result is clinically impactful because methylphenidate is considered as the first-line pharmacotherapy for ADHD, with approximately 40 million prescriptions dispensed every year (Schubert et al., 2010). This result could potentially explain why many patients have an unsatisfactory response to the treatment. Further clinical studies in patients with ADHD with larger sample sizes are needed to fully understand the effect of CES1 variants on the efficacy and toxicity of methylphenidate, and how methylphenidate doses should be adjusted based on a patient’s CES1 genotypes.
G143E and Clopidogrel (Plavix).
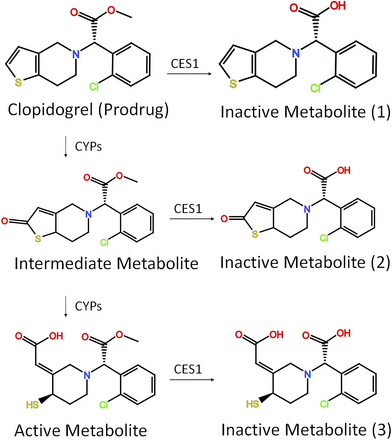
Clopidogrel is a P2Y12 inhibitor and has several clinical indications, including myocardial infarction prophylaxis, cerebrovascular accident prophylaxis, and peripheral arterial occlusive disease prophylaxis. Clopidogrel is usually considered as the first-line antiplatelet agent because of its proven efficacy and cost-effectiveness (Wiviott et al., 2007; Wallentin et al., 2009; Roe et al., 2012). Clopidogrel is a non–ester-prodrug that needs to be activated by two oxidation reactions via several CYPs (Fig. 3). CYP2C19 pharmacogenetics and its impact on clopidogrel activation have been extensively studied. The Clinical Pharmacogenetics Implementation Consortium guidelines and the FDA both recommend intermediate and poor metabolizers of CYP2C19 to use an alternative antiplatelet agent, such as ticagrelor or prasugrel (Scott et al., 2013). Clopidogrel and its intermediate and active metabolites are all CES1 substrates and metabolized by CES1 to inactive hydrolytic metabolites (Fig. 3). Approximately 85% of clopidogrel is hydrolyzed by CES1, and only 15% clopidogrel enters the CYPs-mediated activation pathway (Zhu et al., 2013). Thus, patients with CES1 dysfunction would have a higher concentration of clopidogrel active metabolite compared with normal CES1 metabolizers when taking the same dose. However, the impact of CES1 on the PK and PD of clopidogrel is less studied than the impacts of CYPs.
Clopidogrel metabolic pathway. Clopidogrel is a non-ester-prodrug that needs to be activated by two oxidation reactions via CYPs. Clopidogrel and its intermediate and active metabolites are all metabolized (deactivated) by CES1.
Two clinical trials support that CES1 G143E carriers have significantly higher plasma concentrations of clopidogrel active metabolite compared with noncarriers. A retrospective subanalysis was performed on participants of the Pharmacogenomics of Antiplatelet Intervention (PAPI) Study (n = 506) and on patients who were treated with clopidogrel at Sinai Hospital (n = 350) to examine the effect of CES1 G143E on clopidogrel metabolism. Study participants received a 300-mg loading dose of clopidogrel followed by a 75-mg maintenance dose for 6 days, and platelet aggregation was measured as a PD marker. A 50% higher active metabolite concentration was observed in G143E carriers (n = 7, 30.3 ng/ml) compared with noncarriers (n = 499, 19.0 ng/ml) (P = 0.001). In addition, the inhibition of adenosine diphosphate (ADP)-induced platelet aggregation was 24% higher in G143E carriers (reduced to 71% from baseline) relative to noncarriers (reduced to 57% from baseline) (P = 0.003) (Lewis et al., 2013; Bozzi et al., 2016; Jiang et al., 2016). Another prospective, single-dose, healthy volunteer (n = 22) clinical study was conducted by Tarkiainen et al. (2015a) to determine the effect of CES1 G143E on clopidogrel metabolism. The authors found that the AUC0–∞ ratio of clopidogrel carboxylic acid [inactive metabolite (1) in Fig. 3] to clopidogrel was 53% less in G143E carriers (n = 10) than noncarriers (n = 12) (P = 0.009). The G143E carriers also exhibited significantly higher plasma concentrations of the parent compound clopidogrel (P = 0.004) and its active metabolite (P = 0.009) compared with noncarriers. In agreement with the PK findings, the average inhibition of P2Y12-mediated platelet aggregation in the carriers was 19% points higher than in noncarriers (P = 0.036) (Zhu et al., 2013; Tarkiainen et al., 2015a). The findings of the above two studies are especially important for patients on triple antithrombotic therapy with a high bleeding risk (Mehta et al., 2001; Steinhubl et al., 2002; Shmyr et al., 2017). Clopidogrel dose adjustment may be necessary to prevent potential toxicity (i.e., bleeding) in patients with CES1 dysfunction.
G143E and Angiotensin-Converting Enzyme Inhibitors.
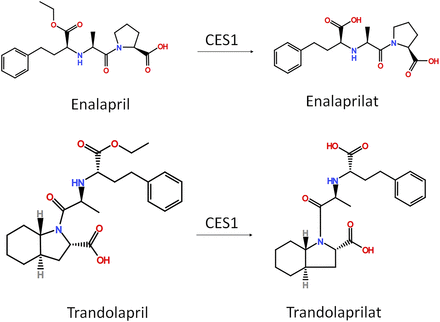
Angiotensin-converting enzyme inhibitors (ACEIs) are generally considered to be the first-line therapy for heart failure and hypertension, and approximately 150 million ACEI prescriptions are filled in the United States annually (Mahmoudpour et al., 2015). Currently, 8 out of 10 FDA-approved ACEIs are ester-containing prodrugs, and all ACEI prodrugs need to be activated by CES1 to exert their intended therapeutic effects (Chaturvedi, 2004; Yancy et al., 2017). The activation is essential for the pharmacological effects because the active metabolites are 10–1000 times more potent than their prodrug forms (Foye et al., 2013). Therefore, patients with CES1 dysfunction would have a lower concentration of the ACEI active metabolite relative to normal CES1 metabolizers (Fig. 4).
ACE inhibitors (enalapril and trandolapril) metabolism. Enalapril and trandolapril are ester-prodrugs that need to be activated by CES1.
A prospective, single-dose pharmacokinetic clinical study was conducted in healthy volunteers to examine the effect of the G143E variant on the activation of the ACEI prodrugs enalapril and quinapril. The AUC0–∞ of the enalapril active metabolite enalaprilat was found to be 20% lower in the G143E carriers (n = 10) than in noncarriers (n = 12) (P = 0.049) (Tarkiainen et al., 2015b). This finding is consistent with an in vitro study that showed that enalapril activation was impaired in liver samples carrying the G143E variant (Wang et al., 2016b). However, the AUCs0-∞ of the quinapril and its active metabolite (quinaprilat) were not significantly different between carriers and noncarriers (P = 0.114). Further investigations are warranted to fully understand the effect of CES1 variants on the PK and PD of ACEI prodrugs.
G143E and Oseltamivir (Tamiflu).
Oseltamivir is an antiviral drug that has an FDA indication for influenza types A and B infections. Even though oseltamivir is rarely effective because of its specific administration requirement (i.e., this medication should be taken within 2 days of onset of symptoms to reduce flu duration by approximately 1 day), oseltamivir remains one of the most prescribed drug products because of flu epidemics (Singh et al., 2003; Dahlgren et al., 2018). As an ester-prodrug, oseltamivir needs to be activated by CES1 into its active metabolite, oseltamivir carboxylate (Shi et al., 2006). An in vitro study based on cell lines stably transfected with CES1 variants suggested the G143E SNP markedly impaired CES1 activity in oseltamivir activation (Zhu and Markowitz, 2009).
To examine the effect of G143E on oseltamivir PK and activation, a prospective, single-dose pharmacokinetic clinical study was conducted in healthy volunteers consisting of nine G143E heterozygotes, one G143E homozygote, and 12 noncarriers. The AUC0–∞ ratio of oseltamivir carboxylate (active metabolite) to oseltamivir (parent molecule) was 23% lower in G143E heterozygotes compared with noncarriers (P = 0.006). The one G143E homozygous individual had an AUC0–∞ of oseltamivir that was approximately 360% greater than that of the noncarriers, indicating that loss of CES1 activity could profoundly impair oseltamivir activation (Tarkiainen et al., 2012).
G143E and Dabigatran and Sacubitril.
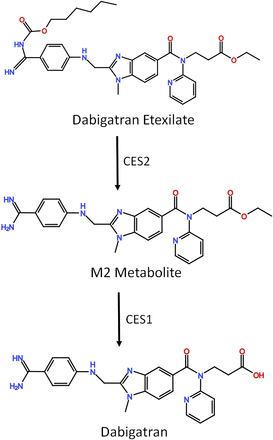
Dabigatran and sacubitril are both prodrugs that need to be activated by CES1 in the liver (Fig. 5). In vitro studies showed that the formation rates of the active metabolites of dabigatran and sacubitril were significantly lower in human livers carrying the G143E variant than in noncarrier samples (Shi et al., 2016b,c). However, it remains undetermined whether the variant can affect the activation and therapeutic response of these two drugs in patients.
Dabigatran metabolic pathway. Dabigatran is a prodrug that activated by both CES1 and CES2.
Pharmacogenetics of Other CES1 Genetic Variants
In addition to G143E, many other CES1 variants have been studied for their effects on the PK and PD of CES1 substrate drugs. However, the results were generally inconclusive, and further studies are needed to determine the clinical significance of these variants.
E220G (rs200707504).
A nonsynonymous variant E220G, commonly referred to as c.662A>G, was suggested to decrease CES1 enzymatic activity in an in silico analysis (Oh et al., 2017). In agreement with that prediction, an in vitro study on transfected cell lines found E220G markedly decreased CES1 activity and the metabolisms of several CES1 substrates, including enalapril, clopidogrel, and sacubitril (Wang et al., 2017). Notably, E220G has a MAF of 0.55% in East Asians but is rare in other populations. To determine the clinical impact of E220G on the PK of a CES1 substrate, a single-dose oseltamivir (75 mg) PK study was conducted in 20 healthy Korean volunteers. In this study, the variant was observed to have a marginal effect on the PK of oseltamivir and its active metabolite (oseltamivir carboxylate); however, the differences were statistically insignificant. In the E220G carriers (n = 8), the AUC0–48 h of oseltamivir was increased by 10% (P = 0.334), and the AUC0–48 h of oseltamivir carboxylate was decreased by 5% (P = 0.513) relative to the noncarriers (n = 12) (Oh et al., 2017).
S75N (rs2307240).
S75N is one of the most common CES1 nonsynonymous SNPs, with MAFs ranging from 2% to 7% in different populations. A retrospective pharmacodynamics analysis was conducted to examine the effect of CES1 S75N on the outcome of clopidogrel therapy in patients with the coronary syndrome (n = 851). The result showed that CES1 S75N carriers (n = 372) had higher incidence of cerebrovascular events (P < 0.001), acute myocardial infarction (P < 0.001), and unstable angina (P < 0.001) compared with noncarriers. The study also found that the S75N polymorphism was more frequent in patients with acute coronary syndrome (MAF 22%) than in the general population (MAF 5%). The authors concluded that there was a significant association between the S75N polymorphism and the outcome of clopidogrel therapy (Xiao et al., 2017). However, this result conflicts with another study that found the S75N variant to be not associated with the outcomes of patients treated with methylphenidate (Johnson et al., 2013). Furthermore, an in vitro study showed the S75N variant did not significantly alter the expression and activity of CES1 in transfected cells and human livers (Wang et al., 2017).
-816A>C (rs3785161).
The -816A>C polymorphism is located in the promoter region of CES1P1 VAR and has been suggested as a potential upregulator of CES1P1 VAR expression (Yoshimura et al., 2008). A prospective clinical study was conducted to examine the impact of -816A>C on the outcome of the ACEI prodrug (imidapril) therapy in patients with hypertension (n = 105). The study found that after 8 weeks of imidapril therapy, -816A>C homozygotes and heterozygotes (n = 47) had greater systolic blood pressure reduction (24.1 mm Hg) compared with noncarriers (17.6 mm Hg) (P = 0.0184), indicating increased CES1 functionality in the carriers. The follow-up in vitro study claimed that the -816A>C SNP may have enhanced transcription of the CES1P1 VAR gene (Geshi et al., 2005). The -816A>C SNP was also evaluated for its impact on the outcomes of dual antiplatelet therapy (i.e., aspirin and clopidogrel) in patients with coronary heart diseases (n = 162). The -816A>C carriers (n = 75) had decreased vasodilator-stimulated phosphoprotein-platelet reactivity index (VASP-PRI) (P = 0.014), indicating increased CES1 function in the carriers (Xie et al., 2014).
However, conflicting findings were reported by other studies. In a study involving the outcome of clopidogrel treatment in patients undergoing percutaneous coronary intervention, -816A>C carriers showed a lower ADP-induced maximum platelet aggregation (21.5%, n = 125) compared with noncarriers (31.7%, n = 124) (P = 0.001), indicating decreased CES1 function (Zou et al., 2014). Zhu et al. (2014) also performed a retrospective pharmacogenetic analysis of the INternational VErapamil SR Trandolapril study (n = 486) and did not find an association between -816A>C and the blood pressure–lowering effect of trandolapril. The follow-up in vitro study also showed -816A>C genotype was not significantly associated with CES1 protein expression and trandolapril activation in human liver samples (n = 100) (Zhu et al., 2016). Other researchers also noted that the CES1P1 VAR gene, which contains -816A>C, is considered functionally insignificant because of its low transcription efficiency (Tanimoto et al., 2007; Hosokawa et al., 2008).
-75G>T (rs3815583).
The -75G>T SNP is located in the promoter region of CES1 and was suspected to alter CES1 expression in the liver; however, the findings are conflicted. A study was performed to determine the association between the variant and appetite reduction (a side effect of methylphenidate) in children with ADHD (n = 213). Appetite reduction was measured by the Barkley Stimulant Side Effect Rating Scale, and methylphenidate dose was titrated up for 3 months as tolerable. The carrier group (n = 129) had worse appetite reduction compared with noncarriers (n = 76) (41% vs. 77%, P = 0.01), indicating that the variant was associated with decreased CES1 function (Bruxel et al., 2013). A study in patients treated with irinotecan, however, showed a contrary finding, suggesting that the -75G>T variant confers greater CES1 function (Sai et al., 2010). CES1 is involved in the conversion of the prodrug irinotecan to its active metabolite, SN-38, and then is further metabolized by UGT1As to inactive SN-38G. Following irinotecan treatment, patients who carried the T allele of this variant had higher plasma (SN-38 + SN-38G)/irinotecan AUC ratios relative to noncarriers (P = 0.027) following irinotecan treatment (Sai et al., 2010).
Other CES1 substrates, isoniazid, and ACEI prodrugs were also studied in the context of -75G>T; however, no significant relationships were found between the variant and the medication responses. In one such study, the variant was evaluated for its effect on the outcomes of ACEI prodrugs in patients with congestive heart failure (n = 200) who underwent ACEI prodrug dose titrations. The study reported -75G>T did not significantly impact plasma angiotensin (AT) II/ATI ratios, and furthermore, the -75G>T variant was not significantly associated with fatal outcomes (i.e., cardiovascular death and all-cause death) (Nelveg-Kristensen et al., 2016). The study with isoniazid had similar results showing no significant association between the variant and isoniazid-induced hepatotoxicity (n = 170) (Yamada et al., 2010).
1168-33C>A (rs2244613).
Dabigatran (Pradaxa) is a prodrug that needs to be activated by both CES1 and CES2 to exert its anticoagulant effect (Fig. 5). Paré and associates (2013) conducted a genome-wide association study of dabigatran in participants (n = 2944) of the Randomized Evaluation of Long-term Anticoagulation Therapy clinical trial. The researchers concluded the CES1 intronic variant 1168-33C>A (rs2244613) is associated with lower trough concentrations of the active metabolite [15% decrease per allele; 95% confidence interval (CI) 10%–19%] and a lower risk of any bleeding (odds ratio, 0.67; 95% CI 0.55–0.82) compared with noncarriers (Paré et al., 2013). However, an in vitro study did not find the variant to be associated with CES1 protein expression and dabigatran metabolism in human livers (Shi et al., 2016b). A prospective study also examined the impact of 1168-33C>A in patients with ADHD that were treated with methylphenidate. The study found the variant to be associated with the occurrence of sadness, a side effect of short-acting methylphenidate. However, researchers concluded this might be due to linkage disequilibrium with two SNPs of the noradrenaline transporter gene (Johnson et al., 2013).
Copy Number Variation (i.e., CES1P1/CES1P1 VAR).
Many researchers have studied the impact of copy number variations (CNVs) on CES1 functionality; however, the results are conflicted. Stage et al. (2017a) found that participants with four functional copies of CES1 (n = 5) had an increased AUC of d‐methylphenidate relative to the control group with two functional copies of CES1 (n = 17) (61% increase, P = 0.011); participants with three copies of CES1 (n = 2) had 45% increased AUC compared with the control group (P = 0.028). Stage et al. (2017b) conducted a similar study with enalapril (n = 43); however, they could not find a statistically significant correlation between CNV and enalapril PK. When Sai et al. (2010) examined the effect of CNV on the irinotecan exposure, they found patients with multiple CES1 copies (i.e., three or four) to have 1.24-fold higher irinotecan AUC relative to patients with two copies of CES1 (P = 0.0134). Many researchers, however, did not find the relationship between CNVs and CES1 function. Suzaki et al. (2013b) evaluated the relationship between CNVs of CES1 and oseltamivir PK parameters but did not find any correlation. Nelveg-Kristensen et al. (2016) studied the relationship between CNV and ACEI prodrugs, and again, no association was found. Moreover, an in vitro study showed CES1 protein expression levels to be comparable among human livers with different copy numbers of functional CES1 gene (Wang et al., 2016b).
Other CES1 SNPs.
In addition to the polymorphisms discussed above, sporadic reports have stated several CES1 SNPs to be associated with the outcomes of CES1 substrate medications. For example, the SNP 1315 + 2025A>C (rs8192950) was associated with a decreased risk of ischemic events in patients (n = 64) having symptomatic extracranial or intracranial stenosis and receiving dual antiplatelet therapy with clopidogrel for a minimum of 5 days (Zhao et al., 2016). Another retrospective subanalysis of a capecitabine clinical study identified associations of 1168-41C>T (rs2244614), 690 + 129del (rs3217164), 95346T>C (rs7187684), -1232A>G (rs1186118) with severe early onset of capecitabine-induced toxicity (Hamzic et al., 2017). None of these findings have been validated independently.
A rare LOF variant, D260fs (c.70DelT), was reported in a clinical study (Zhu et al., 2008). D260fs causes a deletion in exon 6, resulting in a frameshift and premature truncation. Moreover, an in vitro study with CES1 variant–transfected cell lines examined the SNPs proximate to the CES1 active site and identified four LOF nonsynonymous SNPs: G142E, G147C, Y170D, and R171C. However, these variants appear to be clinically insignificant because of their low MAFs (<0.4%) (Wang et al., 2017).
The above-mentioned CES1 SNPs and their impacts on the PK and PD of CES1 substrate medications are summarized in Table 3.
CES1 SNPs and their impacts on the PK and PD of CES1 substrate medications
Nongenetic Factors Affecting CES1 Expression and Activity
Developmental Expression of CES1
The developmental expression patterns of CES1 in human and mouse livers were similar, and many in vitro studies have suggested that hepatic CES1 protein expression increases with age (Zhu et al., 2009a; Hines et al., 2016; Boberg et al., 2017). An in vitro study with human liver samples (n = 104) demonstrated the adult group (≥18 years of age) to have had higher CES1 expression than children (0 days–10 years); meanwhile, child group had higher CES1 expression than fetuses (82–224 gestation days). A follow-up study with liver microsomes showed that, in parallel with expression level, CES1 activity on hydrolyzing its substrate oseltamivir was also positively correlated with age (Yang et al., 2009). The same group did a similar in vitro human liver study with a slightly different age bracket, in which the liver samples were divided into five age groups: 1–31 days old (group 1), 35–70 days old (group 2), 89–119 days old (group 3), 123–198 days old (group 4), and over 18 years old (group 5). Neonates (group 1) had 10% of the CES1 expression and hydrolysis levels compared with the adult group (group 5); pediatric groups (Group 2–4) had approximately 50% of the CES1 expression and hydrolysis levels compared with an adult (Shi et al., 2011). Lastly, a similar in vitro study quantified CES1 protein levels in human liver samples of various ages (n = 165). CES1 expression levels were 4.76 pmol/mg from birth to 3 weeks (n = 36); 15.8 pmol/mg for those aged 3 weeks to 6 years (n = 90); and 16.6 pmol/mg for ages 6–18 years (n = 36). The study team concluded that the median CES1 expression level is directly correlated with age (P < 0.001) (Hines et al., 2016). Overall, CES1 expression and activity levels are lower in neonates and pediatric cohorts; further studies are warranted to investigate the potential effect of CES1 maturation on the treatment outcome of CES1 substrate medications in patients in the early stages of development.
Sex Difference of CES1 Expression
Both in vitro and clinical studies have suggested that CES1 expression is higher in females than in males (Patrick et al., 2007; Zhu et al., 2009a; Shi et al., 2016b). A PK study on healthy volunteers revealed that males had significantly higher exposure to d-methylphenidate than females (Patrick et al., 2007). Nonetheless, females experienced a more pronounced stimulant effect despite their lower exposure. Shi et al. (2016b) observed significantly higher CES1 activity in female human liver samples (n = 56) compared with male samples (n = 46). A follow-up in vitro study with dabigatran suggested CES1 activity was higher in females than males (Shi et al., 2016b). However, such difference was not observed in another in vitro study using human liver samples (n = 32) and mouse liver samples (n = 9) (Zhu et al., 2009a). Further study is needed to examine the impact of sex on the CES1 expression level and the PK and PD of CES1 substrates.
Drug-Drug Interactions
CES1 Inhibitor—Alcohol.
To date, ethanol is the only known CES1 inhibitor that has been confirmed in multiple in vivo and in vitro studies. The impact of ethanol on the metabolism of the CES1 substrate, methylphenidate, was tested in healthy volunteers (n = 14) (Zhu et al., 2017). D-methylphenidate comes as a single active ingredient (Focalin) or in combination with l-methylphenidate (racemic mixture, Ritalin). D-methylphenidate is approximately 10 times more pharmacologically potent than l-methylphenidate, whereas l-methylphenidate is a more efficient CES1 substrate (Fig. 2). This clinical study used a pulsatile dosing regimen with methylphenidate (dl-methylphenidate 40 mg or d-methylphenidate 20 mg) and ethanol (0.6 g/kg, 4 hours after methylphenidate dose) to eliminate any potential confounding effect of ethanol on methylphenidate absorption because the methylphenidate drug products (i.e., Ritalin and Focalin) might undergo faster gastric dissolution in the stomach if administered with alcohol. When alcohol and d-methylphenidate (Focalin) were coadministered, the Cmax of d-methylphenidate was elevated by 27% (P = 0.001), and the AUC4→8 h was elevated by 20% (P < 0.01); when alcohol and dl-methylphenidate (Ritalin) were coadministered, the Cmax of d-methylphenidate was elevated by 35% (P < 0.01), and the AUC4→8 h was elevated by 25% (P < 0.05) (Zhu et al., 2017). These results are consistent with the previous clinical trial by Patrick et al. (2013). In that study, when alcohol and d-methylphenidate (Focalin) were coadministered, the d-methylphenidate AUC was increased by 14%; when alcohol and dl-methylphenidate (Ritalin) were coadministered, the d-methylphenidate AUC was increased by 21% (Patrick et al., 2013). Patrick and colleagues (2007) also showed that the coadministration of alcohol 30 minutes before or 30 minutes after methylphenidate had a similar impact on methylphenidate exposure. Both authors concluded that alcohol is a strong inhibitor of CES1, and the impact of CES1 inhibition is greater for dl-methylphenidate (Ritalin) than for d-methylphenidate (Focalin). Additionally, the DDI between methylphenidate and ethanol produced the transesterification metabolites d-ethylphenidate and l-ethylphenidate, and the plasma concentrations of l-ethylphenidate were much higher than d-ethylphenidate because of l-ethylphehidate being a more efficient CES1 substrate (Zhu et al., 2011, 2017). Other in vivo studies with mice demonstrated similar results (Griffin et al., 2010, 2013; Bell et al., 2011b).
The impact of alcohol on the CES1 function was also examined in the context of a different CES1 substrate, oseltamivir. A prospective health volunteer PK study (n = 18) examined the interaction between oseltamivir 150 mg (a recommended daily dose for the treatment of influenza) and alcohol. Alcohol increased the oseltamivir AUC0–6 h by 27% (P = 0.011) and decreased the AUC0–6 h ratio of the active metabolite oseltamivir carboxylate to the parent compound oseltamivir by 34% (P < 0.001) (Parker et al., 2015). However, coadministration of alcohol did not significantly affect the AUC0–24 h of oseltamivir carboxylate. These results are consistent with in silico analysis of the DDI between alcohol and oseltamivir (Hu et al., 2014).
Other CES1 Inhibitors: Cannabis, Protease Inhibitors, Aripiprazole, Isradipine, Tacrolimus, Valproate.
Besides alcohol, many drug products on the market have been suggested to be potent inhibitors of CES1 mainly by in vitro investigations (Table 4). A further clinical study with a validated CES1 substrate is needed to determine the clinical significance of these CES1 inhibitors.
Drug-drug interaction summary
An in vitro study with CES1-transfected cells suggested that cannabis [i.e., tetrahydrocannabinol (THC), cannabidiol (CBD), and cannabinol (CBN)] can act as a potential CES1 inhibitor. The inhibition constant (Ki) values for THC, CBD, and CBN were 0.541, 0.974, and 0.263 µM (0.170, 0.306, and 0.0817 µg/ml), respectively (Qian et al., 2019). This result could be clinically impactful because the use of cannabis is expected to increase in the next few years (Hasin, 2018).
Several protease inhibitors (i.e., nelfinavir, amprenavir, atazanavir, ritonavir, and saquinavir) were identified as CES1 inhibitors by an in silico analysis and later confirmed by an in vitro incubation study. Among those, nelfinavir had a significantly higher inhibitory effect than the other agents. The relative CES1 activity toward p-nitrophenyl acetate (PNPA) (a CES1 substrate) was 5.2%, 74.2%, 51.7%, 76.9%, and 67.8% of the control after incubation with nelfinavir, ritonavir, amprenavir, saquinavir, and atazanavir, respectively (Rhoades et al., 2012).
An in vitro study suggested aripiprazole, perphenazine, thioridazine, and fluoxetine to be potent inhibitors of CES1, and a complementary animal study (n = 10) with FVB mice demonstrated that coadministration of aripiprazole and methylphenidate (CES1 substrate) significantly increased the plasma concentrations of dl-methylphenidate (P < 0.01) (Zhu et al., 2010).
Moreover, a total of 27 cardiovascular, antiplatelet, anticoagulant, and immunosuppressant drugs have been tested for CES1 inhibition using human liver microsomes and recombinant CES1. The results suggested isradipine (a dihydropyridine calcium antagonist) and tacrolimus (an immunosuppressive agent) to be potent CES1 inhibitors. CES1 activity toward PNPA was decreased to 17.6% with isradipine and 28.4% with tacrolimus (Thomsen et al., 2014).
An in vitro study suggested valproate could inhibit CES1 function and affect rufinamide metabolism in both microsomes and cytosol. This result could be clinically significant because the two antiepileptic medications are often prescribed together when monotherapy is ineffective (Williams et al., 2011).
A combined ensemble docking and machine learning approach was used to identify potential CES1 inhibitors from 1114 FDA-approved drugs. Among the identified inhibitor candidates, four drugs including diltiazem, benztropine, iloprost, and treprostinil were found to inhibit CES1 activity in vitro with IC50 values ranging from 13.9 to 391.6 µM (Briand et al., 2019).
Lastly, an in vitro study suggested that some naturally occurring oxysterols and fatty acids might significantly inhibit CES1 activity with IC50 values within the micromolar range (Crow et al., 2010). These compounds could potentially affect CES1-mediated detoxification and drug metabolism in vivo.
CES1 Inducers.
Overall, CES1 inducers are understudied relative to its inhibitors. Evidence suggests that various nuclear receptors might be involved in the regulation of CES1 expression (Staudinger et al., 2010). For example, several agonists of peroxisome proliferator-activated receptors induced the mRNA expressions of several CES1 isoforms in mouse livers (Jones et al., 2013). A moderate increase of CES1 expression was observed in human hepatocytes treated with rifampicin, a prototypical human pregnane X receptor–activating agent (Shi et al., 2008). An in vivo study with mice suggested that glucose could induce hepatic CES1 expression by stimulating CES1 promoter activity and increasing acetylation of histone 3 and histone 4 in the CES1 chromatin, indicating a potential role of CES1 in glucose homeostasis (Xu et al., 2014). Moreover, phenobarbital induced CES1 expression in mouse livers, and the inducibility was more prominent in neonatal mice relative to adult mice (Xiao et al., 2012). Again, a further clinical investigation is needed to determine the impacts of CES1 inducers on the PK and PD of CES1 substrate medications.
Drug-Drug Interactions between CES1 Substrates.
In addition to CES1 inhibitors and inducers, concomitant use of multiple CES1 substrate drugs can theoretically impact the substrate metabolism by competitively inhibiting the CES1. This hypothesis has been tested in several studies. An in vitro study suggested trandolapril and enalapril might increase clopidogrel activation (Kristensen et al., 2014). Consistent with the in vitro study, a follow-up retrospective clinical study reported the concomitant use of ACEI prodrugs and clopidogrel increases the risk of clinically important bleeding in patients with myocardial infarction (n = 70,934) (P = 0.002). The clinical significance of this finding is, however, debatable because the hazard ratio of clinically significant bleeding for patients on concomitant therapy was 1.10 (95% CI 0.97–1.25) (Kristensen et al., 2014). Another clinical study with the similar design did not report a significant association between the composite cardiovascular outcome and the concomitant use of ACEI prodrugs and clopidogrel in patients with myocardial infarction (n = 45,918). The adjusted odds ratios were 0.94 (95% CI 0.76–1.16) for the perindopril and 0.97 (95% CI 0.80–1.18) for ramipril, relative to lisinopril, an ACEI not metabolized by CES1 (Cressman et al., 2015).
Disease States Related to CES1
A prospective clinical study was conducted in monozygotic and dizygotic twin subjects (62–83 years) with (n = 48) or without (n = 247) type 2 diabetes mellitus to examine the association of CES1 with adiposity and metabolic function. CES1 mRNA expression level in adipose tissue was positively associated with body mass index (P < 0.001), fasting glucose level (P = 0.002), insulin (P = 0.006), and triglycerides (P = 0.003) (Friedrichsen et al., 2013). Recent studies have also found that CES1 function was positively correlated with increased liver lipid storage and plasma lipid concentrations, indicating that CES1 might be heavily involved in lipid metabolism and is a potential drug target for the treatment of human metabolic disorders (Kaddurah-Daouk et al., 2018; Lian et al., 2018a,b).
Conclusion and Future Directions
In sum, G143E (rs71647871) is the only clinically significant LOF CES1 variant identified to date, and alcohol is the only potent CES1 inhibitor that significantly affect CES1-mediated drug metabolism both in vivo and in vitro. However, G143E (MAF 2%–4%, carrier frequency 4%–8%) and alcohol-induced DDI are only able to explain a small portion of the interindividual variability in the CES1 function. Previous in vitro studies have demonstrated marked variability of CES1 activity and expression in human liver samples not carrying G143E (Shi et al., 2016a; Wang et al., 2016b). In fact, analysis of the correlation between CES1 expression and activity revealed that the majority of interindividual variability in the CES1 function is due to variation in CES1 protein expression (Wang et al., 2016b).
Unfortunately, the mechanism by which CES1 protein expression is regulated remains largely unexplored. Notably, most of the existing gene expression regulation studies were based upon the measurement of mRNA expression levels. However, increasing evidence suggests that mRNA expression correlates poorly with protein expression for many genes, including CES1 and most DMEs, which could result in false identification of gene expression regulators (Ohtsuki et al., 2012). Recent advances in liquid chromatography tandem mass spectrometry–based proteomics have allowed for accurate CES1 protein quantification. The application of CES1 proteomics in a large set of clinical samples (e.g., human livers) is expected to uncover important factors influencing CES1 expression, such as genetic polymorphisms, disease conditions, inducers, and post-transcriptional modification (Wang et al., 2016a; He et al., 2019); the findings from such research will lead to the development of an individualized pharmacotherapy approach for improving the efficacy and safety of many medications metabolized by CES1.
Authorship Contributions
Wrote or contributed to the writing of the manuscript: Her, Zhu.
Footnotes
- Received October 17, 2019.
- Accepted December 16, 2019.
This work was supported in part by the National Heart, Lung, and Blood Institute [Grant R01HL126969].
The authors state no conflict of interests.
Abbreviations
- AA
- amino acid
- ACEI
- angiotensin-converting enzyme inhibitor
- ADHD
- attention deficit hyperactivity disorder
- ADP
- adenosine diphosphate
- AT
- angiotensin
- AUC
- area under the curve
- BCS
- Biopharmaceutics Classification System
- CBD
- cannabidiol
- CBN
- cannabinol
- CES
- carboxylesterase
- CI
- confidence interval
- CNV
- copy number variation
- CYP
- cytochrome p450
- DABE
- dabigatran etexilate
- DME
- drug-metabolizing enzyme
- FDA
- Food and Drug Administration
- ID
- identification
- LOF
- loss-of-function
- M1
- dabigatran etexilate intermediate metabolite 1
- M2
- dabigatran etexilate intermediate metabolite 2
- MAF
- minor allele frequency
- PAPI
- Pharmacogenomics of Antiplatelet Intervention
- PD
- pharmacodynamics
- PK
- pharmacokinetics
- PNPA
- p-nitrophenyl acetate
- SNP
- single-nucleotide polymorphism
- THC
- tetrahydrocannabinol
- UGT1A
- UDP-glucuronosyltransferase family 1 member A1
- VASP-PRI
- vasodilator-stimulated phosphoprotein-platelet reactivity index
- Copyright © 2020 by The American Society for Pharmacology and Experimental Therapeutics